

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MFLPNDTQFHPSSFLLLGIPGLETLHIWIGFPFCAVYMIALIGNFTILLVIKTDSSLHQPMFYFLAMLATTDVGLSTATIPKMLGIFWINLRGIIFEACLTQMFFIHNFTLMESAVLVAMAYDSYVAICNPLQYSAILTNKVVSVIGLGVFVRALIFVIPSILLILRLPFCGNHVIPHTYCEHMGLAHLSCASIKINIIYGLCAICNLVFDITVIALSYVHILCAVFRLPTHEARLKSLSTCGSHVCVILAFYTPALFSFMTHRFGRNVPRYIHILLANLYVVVPPMLNPVIYGVRTKQIYKCVKKILLQEQGMEKEEYLIHTRF | |
| CCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSHHCCCCCCCCHHHHHHHHHHHHCCCCCCCCSSSCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCC | |
| 9988999871410026899898668899999999999999999999888774688754328999999999999999833999999994799868888999999999999999999999998020662056544603087889999999999999999998999994089999992587221207678883378269799999999999998999999999999998178897688898550699999999999989988210014798881789999999973786646800006569999999999733766642333013689 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MFLPNDTQFHPSSFLLLGIPGLETLHIWIGFPFCAVYMIALIGNFTILLVIKTDSSLHQPMFYFLAMLATTDVGLSTATIPKMLGIFWINLRGIIFEACLTQMFFIHNFTLMESAVLVAMAYDSYVAICNPLQYSAILTNKVVSVIGLGVFVRALIFVIPSILLILRLPFCGNHVIPHTYCEHMGLAHLSCASIKINIIYGLCAICNLVFDITVIALSYVHILCAVFRLPTHEARLKSLSTCGSHVCVILAFYTPALFSFMTHRFGRNVPRYIHILLANLYVVVPPMLNPVIYGVRTKQIYKCVKKILLQEQGMEKEEYLIHTRF | |
| 2444132423031020000012132020001101201220332121000002015100100010001001000000200002000000020440304000000120031023011001100300000002001000000231001000000321201120210102103104520000000102100100023030011000202312331231032011100200020024601320120010010001003231331010010033002100010022100300330030000203400410030014346154431104247 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSHHCCCCCCCCHHHHHHHHHHHHCCCCCCCCSSSCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCC MFLPNDTQFHPSSFLLLGIPGLETLHIWIGFPFCAVYMIALIGNFTILLVIKTDSSLHQPMFYFLAMLATTDVGLSTATIPKMLGIFWINLRGIIFEACLTQMFFIHNFTLMESAVLVAMAYDSYVAICNPLQYSAILTNKVVSVIGLGVFVRALIFVIPSILLILRLPFCGNHVIPHTYCEHMGLAHLSCASIKINIIYGLCAICNLVFDITVIALSYVHILCAVFRLPTHEARLKSLSTCGSHVCVILAFYTPALFSFMTHRFGRNVPRYIHILLANLYVVVPPMLNPVIYGVRTKQIYKCVKKILLQEQGMEKEEYLIHTRF | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.23 | 0.86 | 2.36 | Download | ---------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDV----------VPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH-VLRQ--------- | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.20 | 0.83 | 2.01 | Download | -----------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC--------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLLLNSTVNPIIYALRSKDLRHAFRSMF----------------- | |||||||||||||||||||
| 3 | 5tgzA | 0.19 | 0.20 | 0.84 | 1.78 | Download | MDIECFMVLNP------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID-----------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------- | |||||||||||||||||||
| 4 | 3uon | 0.17 | 0.22 | 0.85 | 1.55 | Download | ----------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQF---------FSNAAVTFGTIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPIPNTV-WTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---------------- | |||||||||||||||||||
| 5 | 3uonA | 0.17 | 0.18 | 0.84 | 1.13 | Download | ----------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFI--VGVRTVEDGECYIQF---------FSNAAVTFGAIAAFYLPVIIMTVLYWHISRASKS----RREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---------------- | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.23 | 0.85 | 2.63 | Download | -----------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDC---SHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ---------- | |||||||||||||||||||
| 7 | 4iaq | 0.17 | 0.21 | 0.81 | 1.69 | Download | ----------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVN--------------------TDHILYTVSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH---L-AIFD-FFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-------------- | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.22 | 0.89 | 3.03 | Download | ----NSD---------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPIV------DIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL----------------- | |||||||||||||||||||
| 9 | 5tgzA | 0.18 | 0.20 | 0.87 | 3.08 | Download | -GGRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG-------NCEKLQSV-----------CSDIHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------- | |||||||||||||||||||
| 10 | 4gpoA | 0.16 | 0.18 | 0.85 | 4.48 | Download | ---------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIIFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
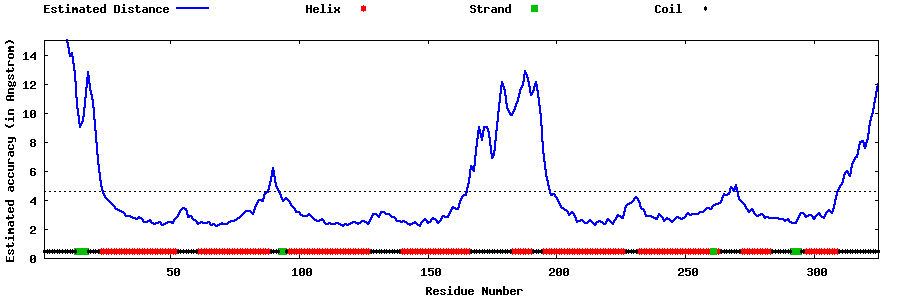
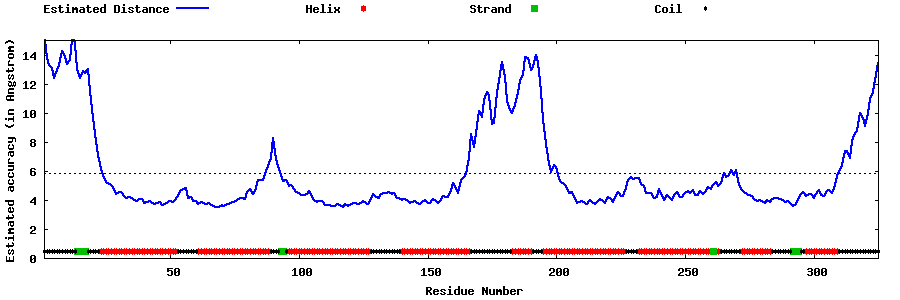
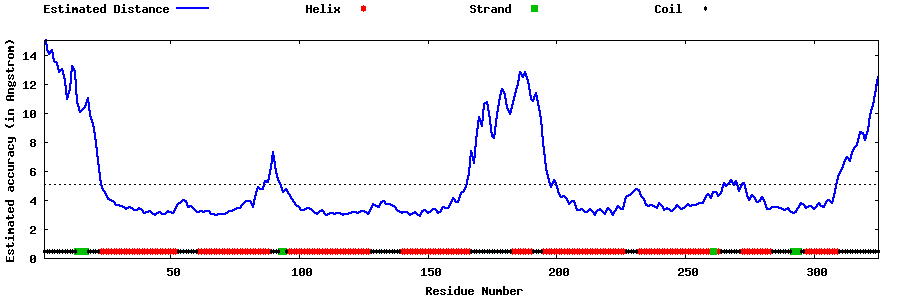
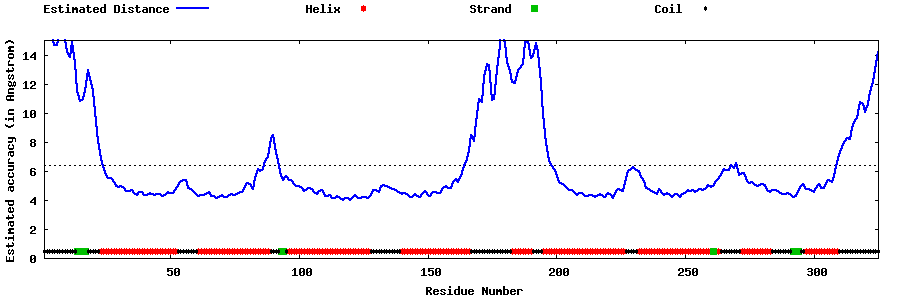
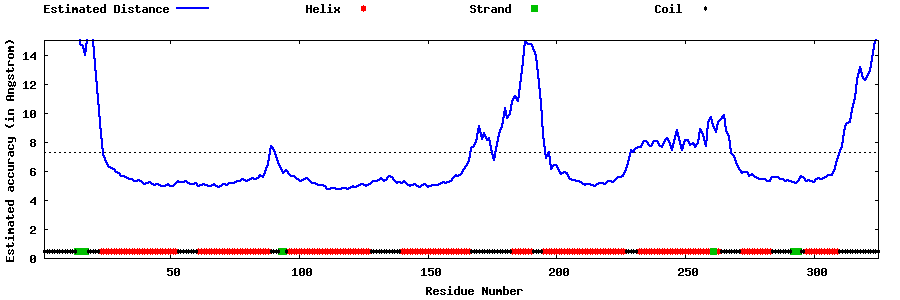
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||