

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNHMSASLKISNSSKFQVSEFILLGFPGIHSWQHWLSLPLALLYLSALAANTLILIIIWQNPSLQQPMYIFLGILCMVDMGLATTIIPKILAIFWFDAKVISLPECFAQIYAIHFFVGMESGILLCMAFDRYVAICHPLRYPSIVTSSLILKATLFMVLRNGLFVTPVPVLAAQRDYCSKNEIEHCLCSNLGVTSLACDDRRPNSICQLVLAWLGMGSDLSLIILSYILILYSVLRLNSAEAAAKALSTCSSHLTLILFFYTIVVVISVTHLTEMKATLIPVLLNVLHNIIPPSLNPTVYALQTKELRAAFQKVLFALTKEIRS | |
| CCCHHHHCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHHCCCCCCC | |
| 941364271889998725001378999987789999999999999999999998777636887543389999999999999998429999999846998788888999999999999999999999980305621566557502888999999999999999999989999933899998947873432066788834782688999999999999999999999999999998078897688898660699999999999989886031677899947899999998427755368011065799999999998845665689 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNHMSASLKISNSSKFQVSEFILLGFPGIHSWQHWLSLPLALLYLSALAANTLILIIIWQNPSLQQPMYIFLGILCMVDMGLATTIIPKILAIFWFDAKVISLPECFAQIYAIHFFVGMESGILLCMAFDRYVAICHPLRYPSIVTSSLILKATLFMVLRNGLFVTPVPVLAAQRDYCSKNEIEHCLCSNLGVTSLACDDRRPNSICQLVLAWLGMGSDLSLIILSYILILYSVLRLNSAEAAAKALSTCSSHLTLILFFYTIVVVISVTHLTEMKATLIPVLLNVLHNIIPPSLNPTVYALQTKELRAAFQKVLFALTKEIRS | |
| 654144403331324231210200000021320200011012012203311210000021141001000200010010000002001020000000203403040000001200210220100000003000000021010000002310010000003212011201100021031045200000001021002000230300111002003323333231032011100200030124601320110010010002003231321010022333011000110121003003300300002034004100300243356358 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHHCCCCCCC MNHMSASLKISNSSKFQVSEFILLGFPGIHSWQHWLSLPLALLYLSALAANTLILIIIWQNPSLQQPMYIFLGILCMVDMGLATTIIPKILAIFWFDAKVISLPECFAQIYAIHFFVGMESGILLCMAFDRYVAICHPLRYPSIVTSSLILKATLFMVLRNGLFVTPVPVLAAQRDYCSKNEIEHCLCSNLGVTSLACDDRRPNSICQLVLAWLGMGSDLSLIILSYILILYSVLRLNSAEAAAKALSTCSSHLTLILFFYTIVVVISVTHLTEMKATLIPVLLNVLHNIIPPSLNPTVYALQTKELRAAFQKVLFALTKEIRS | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.21 | 0.85 | 2.42 | Download | ----------------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFF--CPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH---LRQ | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.22 | 0.83 | 2.08 | Download | ------------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDRMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 3 | 4iaqA | 0.16 | 0.19 | 0.83 | 1.95 | Download | ---------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTG--RWTLVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPP------FFWRQASE---------------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 4 | 4ib4 | 0.18 | 0.23 | 0.86 | 1.54 | Download | --------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKG--IET----NPNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADSNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCQTMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---- | |||||||||||||||||||
| 5 | 3uonA | 0.16 | 0.19 | 0.84 | 1.13 | Download | -----------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQF--IVGVRTVEDGECYIQF---------FSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFIITWAPYNVMVLINFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.21 | 0.85 | 2.60 | Download | ------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC--PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 7 | 4iaq | 0.16 | 0.21 | 0.81 | 1.68 | Download | -------------------------------WKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWLPFFIISLVMPIHLAIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF------- | |||||||||||||||||||
| 8 | 4buoA | 0.14 | 0.19 | 0.87 | 3.18 | Download | -----------NSD---------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGV------------DTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTFYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL--------- | |||||||||||||||||||
| 9 | 5tgzA | 0.22 | 0.22 | 0.87 | 3.09 | Download | --------GGRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSML--STVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 10 | 4gpoA | 0.17 | 0.20 | 0.85 | 4.40 | Download | ----------------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL---------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
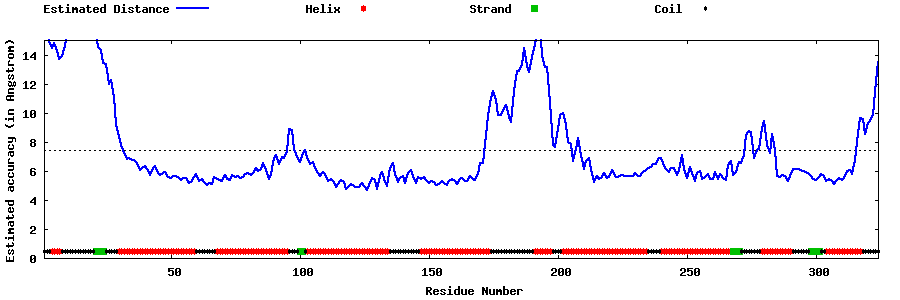
| Generated 3D models | Estimated local accuracy of models | ||
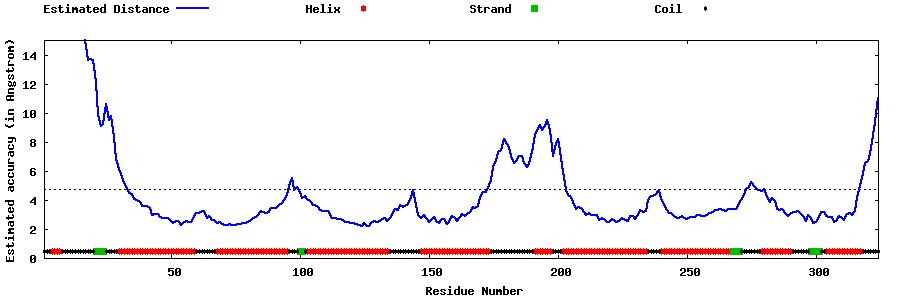 | |||
|
|
|||
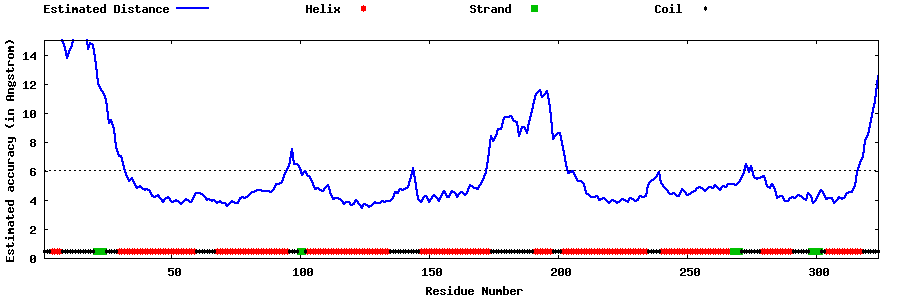 | |||
|
| |||
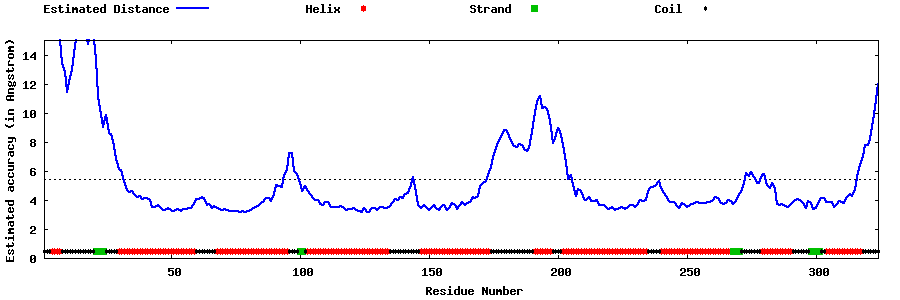 | |||
|
| |||
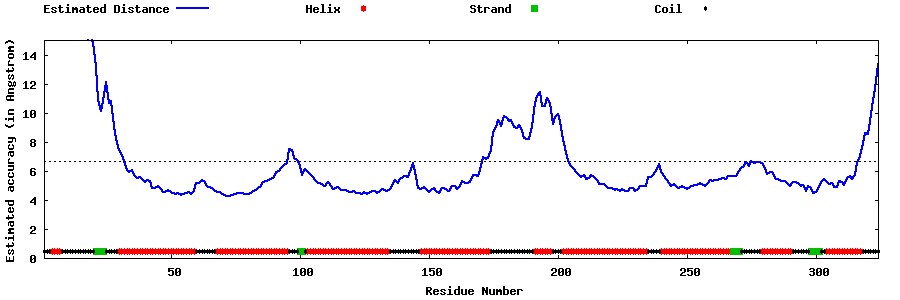 | |||
|
| |||
 | |||
|
| |||