

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MIQPMASPSNSSTVPVSEFLLICFPNFQSWQHWLSLPLSLLFLLAMGANTTLLITIQLEASLHQPLYYLLSLLSLLDIVLCLTVIPKVLAIFWYDLRSISFPACFLQMFIMNSFLPMESCTFMVMAYDRYVAICHPLRYPSIITNQFVAKASVFIVVRNALLTAPIPILTSLLHYCGENVIENCICANLSVSRLSCDNFTLNRIYQFVAGWTLLGSDLFLIFLSYTFILRAVLRFKAEGAAVKALSTCGSHFILILFFSTILLVVVLTNVARKKVPMDILILLNVLHHLIPPALNPIVYGVRTKEIKQGIQKLLQRGR | |
| CCCCCCCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCC | |
| 976445778999872421226799998788999999999999999999999877774788754218999999999999999854999999983699878888899999999999999999999998031785166645761288899999999999999999999999994489999895787444217678883578258899999999999999899999999999999907889778889855169999999999998987712346799788189999999996277663680110657999999999960489 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MIQPMASPSNSSTVPVSEFLLICFPNFQSWQHWLSLPLSLLFLLAMGANTTLLITIQLEASLHQPLYYLLSLLSLLDIVLCLTVIPKVLAIFWYDLRSISFPACFLQMFIMNSFLPMESCTFMVMAYDRYVAICHPLRYPSIITNQFVAKASVFIVVRNALLTAPIPILTSLLHYCGENVIENCICANLSVSRLSCDNFTLNRIYQFVAGWTLLGSDLFLIFLSYTFILRAVLRFKAEGAAVKALSTCGSHFILILFFSTILLVVVLTNVARKKVPMDILILLNVLHHLIPPALNPIVYGVRTKEIKQGIQKLLQRGR | |
| 734402434131423021010000012132020001102201220231121000002014100100020001002201000200102000000021350304000000120021023010000100300000002101000000241001000000321201120110002103104620000000102100200033030011110200332333123103201110020001012560132012001001000200323133101001003300210001001210030033003000020340141004003558 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCC MIQPMASPSNSSTVPVSEFLLICFPNFQSWQHWLSLPLSLLFLLAMGANTTLLITIQLEASLHQPLYYLLSLLSLLDIVLCLTVIPKVLAIFWYDLRSISFPACFLQMFIMNSFLPMESCTFMVMAYDRYVAICHPLRYPSIITNQFVAKASVFIVVRNALLTAPIPILTSLLHYCGENVIENCICANLSVSRLSCDNFTLNRIYQFVAGWTLLGSDLFLIFLSYTFILRAVLRFKAEGAAVKALSTCGSHFILILFFSTILLVVVLTNVARKKVPMDILILLNVLHHLIPPALNPIVYGVRTKEIKQGIQKLLQRGR | |||||||||||||||||||||||||
| 1 | 3emlA | 0.16 | 0.22 | 0.86 | 2.35 | Download | --------------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSRQ | |||||||||||||||||||
| 2 | 5tgzA | 0.17 | 0.21 | 0.85 | 2.00 | Download | ----------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---- | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.22 | 0.85 | 1.93 | Download | -------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTG--RWTLVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLP-------PFFWRQAS--------------ECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI-----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK- | |||||||||||||||||||
| 4 | 3uon | 0.17 | 0.19 | 0.87 | 1.56 | Download | ---------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF------S---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPIPNTV-WTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--- | |||||||||||||||||||
| 5 | 3uonA | 0.17 | 0.15 | 0.86 | 1.14 | Download | ---------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFI--VGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--- | |||||||||||||||||||
| 6 | 3emlA | 0.16 | 0.22 | 0.86 | 2.60 | Download | ----------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFF---CPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHV | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.23 | 0.83 | 1.72 | Download | ----------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWLPFFIISLVMPIHL-AIFD-FFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK- | |||||||||||||||||||
| 8 | 4buoA | 0.18 | 0.21 | 0.87 | 2.92 | Download | ---------NSD---------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGI-----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQRAVVIAFVVCWLPYHVRRLMFCYI--------SDEQWTYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL---- | |||||||||||||||||||
| 9 | 5tgzA | 0.16 | 0.21 | 0.90 | 3.06 | Download | ----GGRGENFMDIEC--FMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---- | |||||||||||||||||||
| 10 | 4gpoA | 0.21 | 0.23 | 0.87 | 4.60 | Download | --------------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL----- | |||||||||||||||||||
| ||||||||||||||||||||||||||
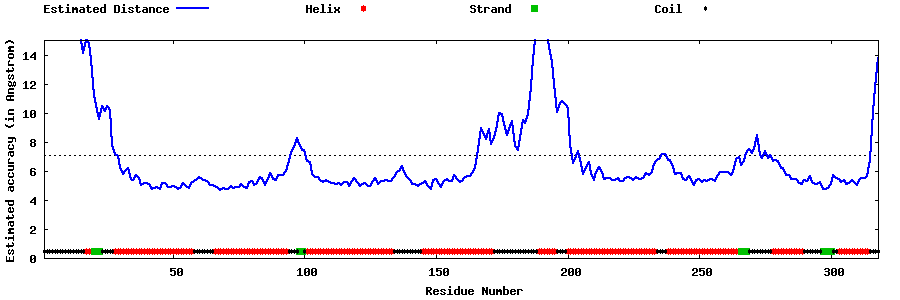
| Generated 3D models | Estimated local accuracy of models | ||
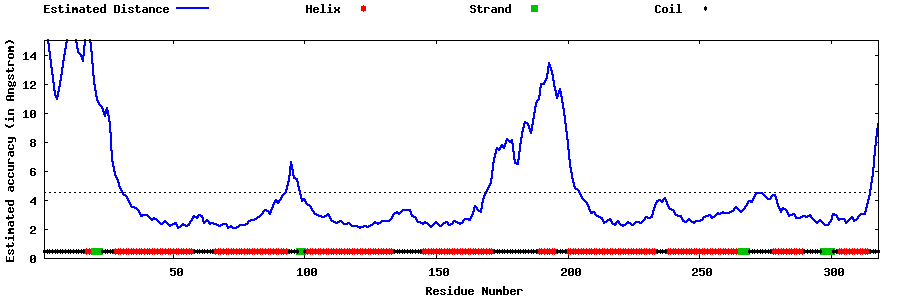 | |||
|
|
|||
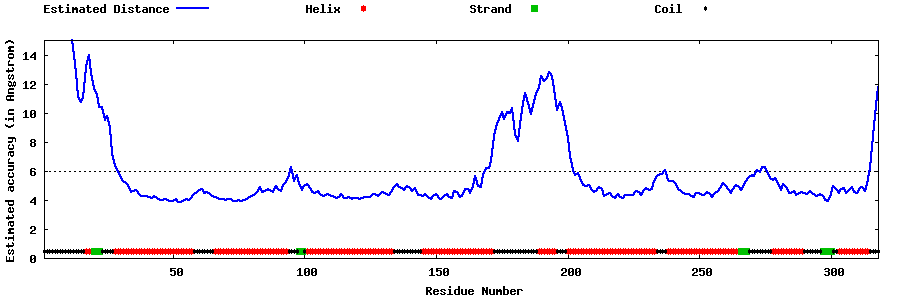 | |||
|
| |||
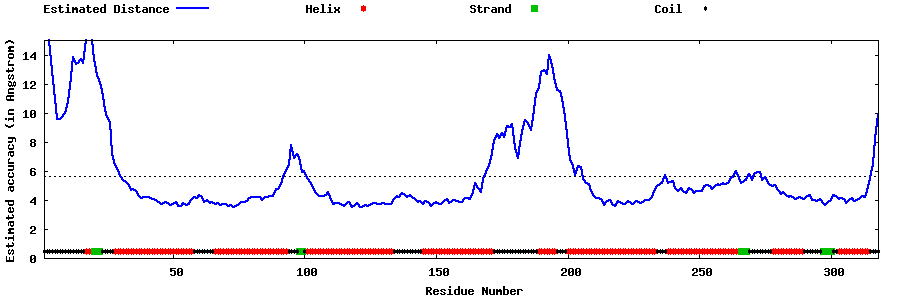 | |||
|
| |||
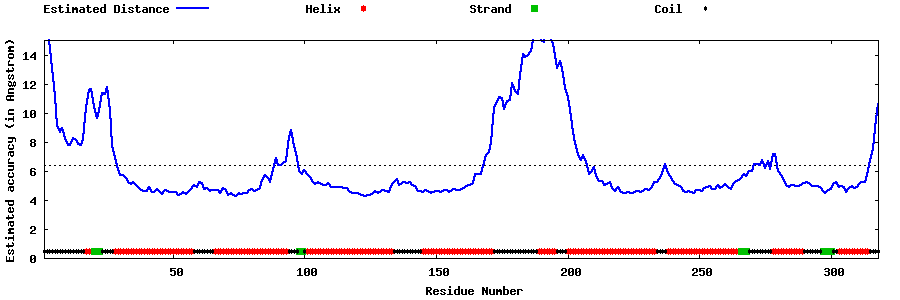 | |||
|
| |||
 | |||
|
| |||