

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGFLSPMHPCRPPTQRRMAAGNHSTVTEFILKGLTKRADLQLPLFLLFLGIYLVTIVGNLGMITLICLNSQLHTPMYYFLSNLSLMDLCYSSVITPKMLVNFVSEKNIISYAGCMSQLYFFLVFVIAECYMLTVMAYDRYVAICHPLLYNIIMSHHTCLLLVAVVYAIGLIGSTIETGLMLKLPYCEHLISHYFCDILPLMKLSCSSTYDVEMTVFFSAGFNIIVTSLTVLVSYTFILSSILGISTTEGRSKAFSTCSSHLAAVGMFYGSTAFMYLKPSTISSLTQENVASVFYTTVIPMLNPLIYSLRNKEVKAAVQKTLRGKLF | |
| CCCCCCCCCCCCCHHHHHCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHCCCC | |
| 97666665346734654105688512034686488982528999999999999999989899999961888777388887779988437202470999998714897885899999999999999999999999975403520622018801678899999999999999999999999766458898567740580888877056873667788889999999999999999999999980366756675422064888997999974340248478998878788789987111142133574405659999999999826679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGFLSPMHPCRPPTQRRMAAGNHSTVTEFILKGLTKRADLQLPLFLLFLGIYLVTIVGNLGMITLICLNSQLHTPMYYFLSNLSLMDLCYSSVITPKMLVNFVSEKNIISYAGCMSQLYFFLVFVIAECYMLTVMAYDRYVAICHPLLYNIIMSHHTCLLLVAVVYAIGLIGSTIETGLMLKLPYCEHLISHYFCDILPLMKLSCSSTYDVEMTVFFSAGFNIIVTSLTVLVSYTFILSSILGISTTEGRSKAFSTCSSHLAAVGMFYGSTAFMYLKPSTISSLTQENVASVFYTTVIPMLNPLIYSLRNKEVKAAVQKTLRGKLF | |
| 52002213334463354035503030110000000432401000023033213212333310020010003000000000200001001000000020002001642201030000001000000000000000000000000021120003003300000011012100200301000003020162100000001200030001313110110012013113303310330331001000103036223101000000210010132012000000335332434200002103200230030000213301300220064347 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCHHHHHCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHCCCC MGFLSPMHPCRPPTQRRMAAGNHSTVTEFILKGLTKRADLQLPLFLLFLGIYLVTIVGNLGMITLICLNSQLHTPMYYFLSNLSLMDLCYSSVITPKMLVNFVSEKNIISYAGCMSQLYFFLVFVIAECYMLTVMAYDRYVAICHPLLYNIIMSHHTCLLLVAVVYAIGLIGSTIETGLMLKLPYCEHLISHYFCDILPLMKLSCSSTYDVEMTVFFSAGFNIIVTSLTVLVSYTFILSSILGISTTEGRSKAFSTCSSHLAAVGMFYGSTAFMYLKPSTISSLTQENVASVFYTTVIPMLNPLIYSLRNKEVKAAVQKTLRGKLF | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.19 | 0.84 | 3.37 | Download | -------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.21 | 0.83 | 1.87 | Download | --------------------ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV-----------------------LPLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------ | |||||||||||||||||||
| 3 | 4iaqA | 0.19 | 0.19 | 0.83 | 2.21 | Download | -----------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL-------PPFFCVVN----------------TDHILYTVYSTVGAFY---FPTLLLIALYGRIYVEARSRIIQLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.21 | 0.86 | 1.55 | Download | ------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE--DVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.21 | 0.92 | 1.26 | Download | FDILVLANEGKVKEAEQLKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHR-NVFF-----IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.19 | 0.84 | 3.55 | Download | --------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 4iaq | 0.21 | 0.21 | 0.81 | 1.73 | Download | --------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQAS---------ECVV----------NTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH-L-AI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 8 | 4ea3A | 0.19 | 0.19 | 0.79 | 2.92 | Download | ------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAIMGSAQVEDIECLVEIPTPQDYWGPVFAICIFLFSF-----------------IVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLG-----VQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR-------- | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.19 | 0.85 | 5.03 | Download | -------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGW-------NNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.20 | 0.87 | 5.61 | Download | ----------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| ||||||||||||||||||||||||||
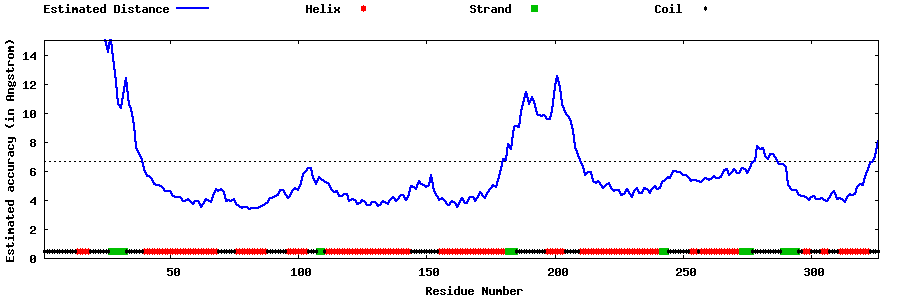
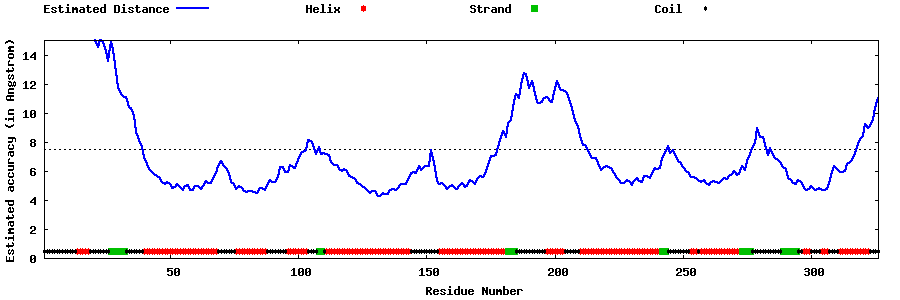
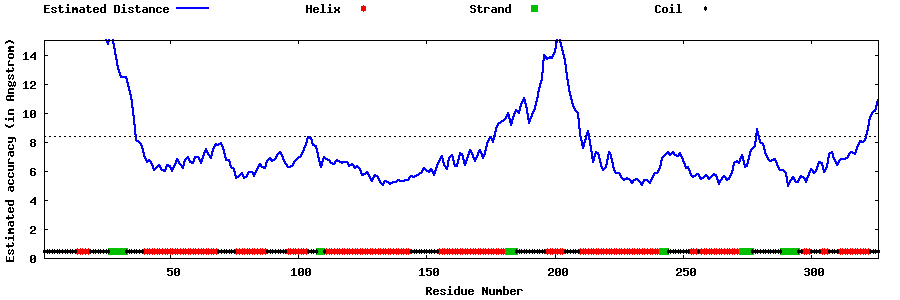
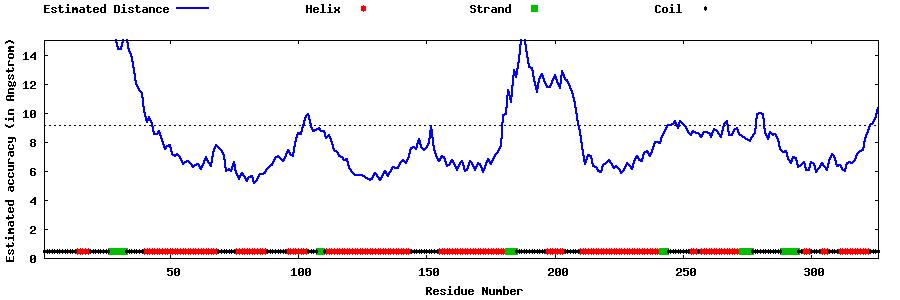
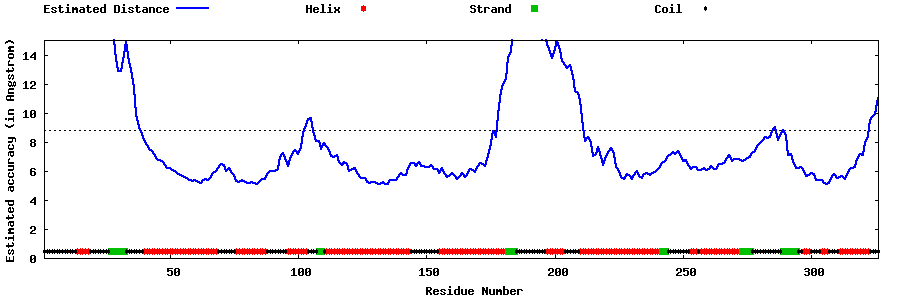
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||