

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MNHVVKHNHTAVTKVTEFILMGITDNPGLQAPLFGLFLIIYLVTVIGNLGMVILTYLDSKLHTPMYFFLRHLSITDLGYSTVIAPKMLVNFIVHKNTISYNWYATQLAFFEIFIISELFILSAMAYDRYVAICKPLLYVIIMAEKVLWVLVIVPYLYSTFVSLFLTIKLFKLSFCGSNIISYFYCDCIPLMSILCSDTNELELIILIFSGCNLLFSLSIVLISYMFILVAILRMNSRKGRYKAFSTCSSHLTVVIMFYGTLLFIYLQPKSSHTLAIDKMASVFYTLLIPMLNPLIYSLRNKEVKDALKRTLTNRFKIPI | |
| CCHHHCCCCCCCCSSHHSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHHHCCCCC | |
| 9445437888885601047863789825289999999999999999887999998528887772888865599873262113529999997048978858999999999999999999999999754134016220288126788999999999999999999999998554188989138632681888777046863767888888899999999999999999999980367757675431065889997999985552137478999888789789987111141233474404649999999999974047889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MNHVVKHNHTAVTKVTEFILMGITDNPGLQAPLFGLFLIIYLVTVIGNLGMVILTYLDSKLHTPMYFFLRHLSITDLGYSTVIAPKMLVNFIVHKNTISYNWYATQLAFFEIFIISELFILSAMAYDRYVAICKPLLYVIIMAEKVLWVLVIVPYLYSTFVSLFLTIKLFKLSFCGSNIISYFYCDCIPLMSILCSDTNELELIILIFSGCNLLFSLSIVLISYMFILVAILRMNSRKGRYKAFSTCSSHLTVVIMFYGTLLFIYLQPKSSHTLAIDKMASVFYTLLIPMLNPLIYSLRNKEVKDALKRTLTNRFKIPI | |
| 5540374133320302000000004324010000230332132123333100200200130000001002000010010000000300010015422000200010000000000000100000000000000211100030033000000120121013103110120030200431100000000200030001323210100010013113303310330331001000103026323101000010210010232011000000424312444200002103200230030000203301400220043317027 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHCCCCCCCCSSHHSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHHHCCCCC MNHVVKHNHTAVTKVTEFILMGITDNPGLQAPLFGLFLIIYLVTVIGNLGMVILTYLDSKLHTPMYFFLRHLSITDLGYSTVIAPKMLVNFIVHKNTISYNWYATQLAFFEIFIISELFILSAMAYDRYVAICKPLLYVIIMAEKVLWVLVIVPYLYSTFVSLFLTIKLFKLSFCGSNIISYFYCDCIPLMSILCSDTNELELIILIFSGCNLLFSLSIVLISYMFILVAILRMNSRKGRYKAFSTCSSHLTVVIMFYGTLLFIYLQPKSSHTLAIDKMASVFYTLLIPMLNPLIYSLRNKEVKDALKRTLTNRFKIPI | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.20 | 0.87 | 3.52 | Download | --------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 2 | 5tgzA | 0.23 | 0.23 | 0.86 | 1.95 | Download | ------GRGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV------------------------LPLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM--------- | |||||||||||||||||||
| 3 | 5tgzA | 0.18 | 0.23 | 0.85 | 2.10 | Download | ----------------GGRGENFMDIPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHV-FHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA-PDQARMDIELAKTLVLILVVLIICWGPLLAIMVVFGKMNKLIKTVFAFCSMLCLLNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 4 | 4djh | 0.12 | 0.21 | 0.88 | 1.54 | Download | -------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------ | |||||||||||||||||||
| 5 | 4yay | 0.19 | 0.24 | 0.91 | 1.27 | Download | VKEAEQLKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIH-RNVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.20 | 0.87 | 3.68 | Download | ---------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 7 | 4iaq | 0.23 | 0.22 | 0.82 | 1.72 | Download | ----------------------------WKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVNT--------------------DHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 8 | 2z73A | 0.16 | 0.21 | 0.96 | 2.96 | Download | -----ETWWYNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDY---------ISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQ | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.20 | 0.87 | 5.32 | Download | --------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 10 | 2ydoA | 0.15 | 0.18 | 0.90 | 5.92 | Download | -----------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
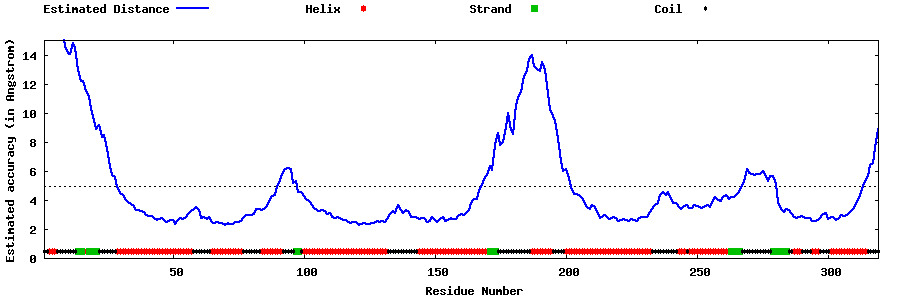 | |||
|
|
|||
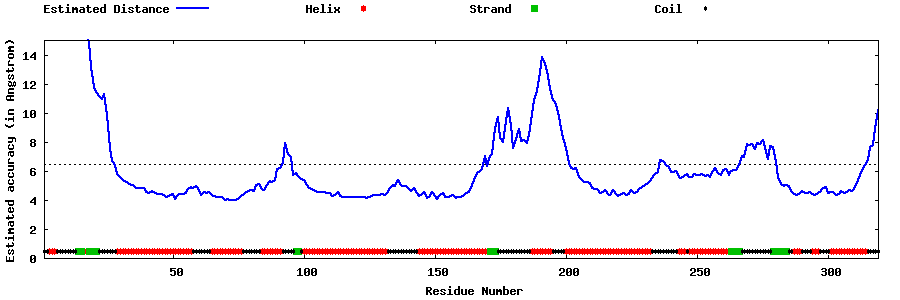 | |||
|
| |||
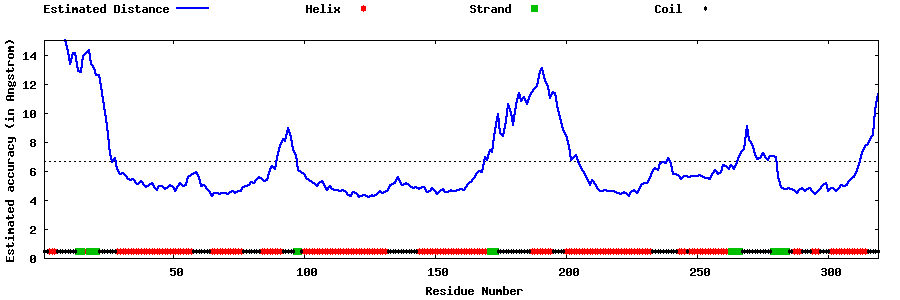 | |||
|
| |||
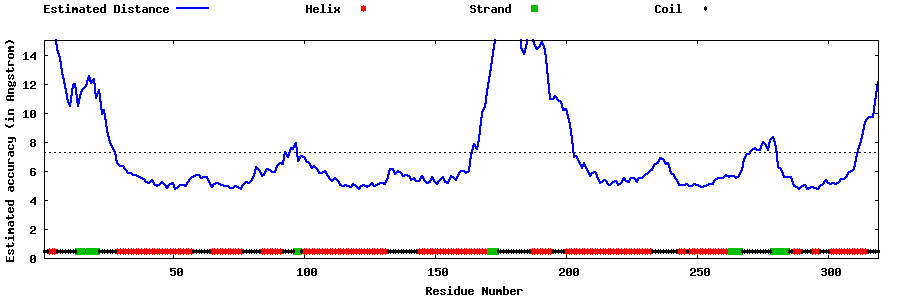 | |||
|
| |||
 | |||
|
| |||