

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MSYSIYKSTVNIPLSHGVVHSFCHNMNCNFMHIFKFVLDFNMKNVTEVTLFVLKGFTDNLELQTIFFFLFLAIYLFTLMGNLGLILVVIRDSQLHKPMYYFLSMLSSVDACYSSVITPNMLVDFTTKNKVISFLGCVAQVFLACSFGTTECFLLAAMAYDRYVAIYNPLLYSVSMSPRVYMPLINASYVAGILHATIHTVATFSLSFCGANEIRRVFCDIPPLLAISYSDTHTNQLLLFYFVGSIELVTILIVLISYGLILLAILKMYSAEGRRKVFSTCGAHLTGVSIYYGTILFMYVRPSSSYASDHDMIVSIFYTIVIPLLNPVIYSLRNKDVKDSMKKMFGKNQVINKVYFHTKK | |
| CCSSHHHHCCCCCCCCCCCCCHHCCCCHHHHHHHHHHHHHHCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHHSSSSSSCC | |
| 95223320477788865301021265246788889999865678851203578658999360899999999999999988879999985088766748888767998844711247088989871489788489999999999999999999999997541340162102880267889999999999999999999999984630789990484415818877661578636677788888999999999999999999999813667476654321558889979998732124583789998787898899882200412435744056498999999999864113003676259 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MSYSIYKSTVNIPLSHGVVHSFCHNMNCNFMHIFKFVLDFNMKNVTEVTLFVLKGFTDNLELQTIFFFLFLAIYLFTLMGNLGLILVVIRDSQLHKPMYYFLSMLSSVDACYSSVITPNMLVDFTTKNKVISFLGCVAQVFLACSFGTTECFLLAAMAYDRYVAIYNPLLYSVSMSPRVYMPLINASYVAGILHATIHTVATFSLSFCGANEIRRVFCDIPPLLAISYSDTHTNQLLLFYFVGSIELVTILIVLISYGLILLAILKMYSAEGRRKVFSTCGAHLTGVSIYYGTILFMYVRPSSSYASDHDMIVSIFYTIVIPLLNPVIYSLRNKDVKDSMKKMFGKNQVINKVYFHTKK | |
| 64232244013111434212210230202113134113403550302001000000043240100001303321321332331001001000300000000010000000100000003000000154320103000100100000000000000000010000001001010400330000000000010010020100000302014312000000031000100012230101100010132133013003302310010001030162231000000002000001220120000003143033341000011021002300200001143014003300332300330203358 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCSSHHHHCCCCCCCCCCCCCHHCCCCHHHHHHHHHHHHHHCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHHSSSSSSCC MSYSIYKSTVNIPLSHGVVHSFCHNMNCNFMHIFKFVLDFNMKNVTEVTLFVLKGFTDNLELQTIFFFLFLAIYLFTLMGNLGLILVVIRDSQLHKPMYYFLSMLSSVDACYSSVITPNMLVDFTTKNKVISFLGCVAQVFLACSFGTTECFLLAAMAYDRYVAIYNPLLYSVSMSPRVYMPLINASYVAGILHATIHTVATFSLSFCGANEIRRVFCDIPPLLAISYSDTHTNQLLLFYFVGSIELVTILIVLISYGLILLAILKMYSAEGRRKVFSTCGAHLTGVSIYYGTILFMYVRPSSSYASDHDMIVSIFYTIVIPLLNPVIYSLRNKDVKDSMKKMFGKNQVINKVYFHTKK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.19 | 0.77 | 3.08 | Download | -----------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL--------RQ | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.23 | 0.76 | 1.76 | Download | ----------------------------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM---------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.20 | 0.76 | 1.90 | Download | ---------------------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPFFWRE---------------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------ | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.20 | 0.78 | 1.53 | Download | ----------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.22 | 0.90 | 1.27 | Download | MRAAALDAQKATPKLSPEMKDFRHGFDILVGKVKEAQEQLKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIH-RNVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--------------- | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.19 | 0.77 | 3.28 | Download | ------------------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-------- | |||||||||||||||||||
| 7 | 4iaq | 0.21 | 0.21 | 0.73 | 1.72 | Download | ------------------------------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVNT-----------------DHIL---YTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------ | |||||||||||||||||||
| 8 | 4ea3A | 0.18 | 0.19 | 0.72 | 2.62 | Download | ----------------------------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEIECLVEIPTPQDYWGPVFAICIFLFSF-----------------IVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLG-----VQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR------------------ | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.19 | 0.77 | 4.55 | Download | -------------------------------------------------------------ISSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-------- | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.20 | 0.82 | 5.16 | Download | --------------------------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFKAAA | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
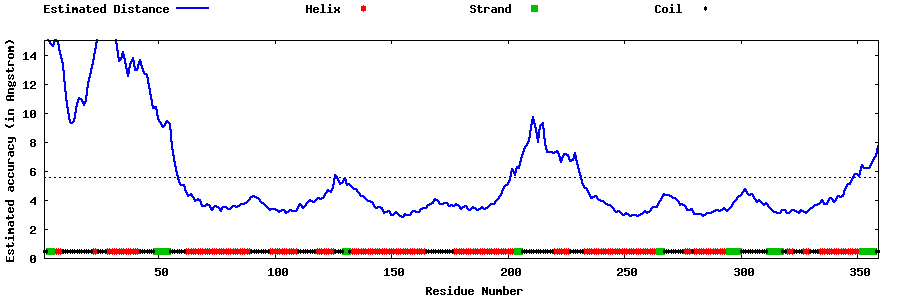 | |||
|
|
|||
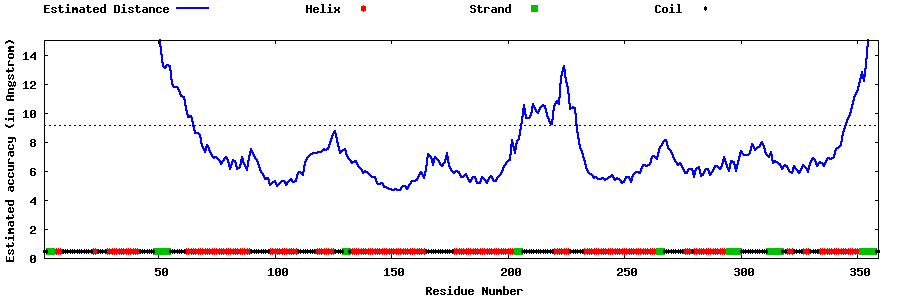 | |||
|
| |||
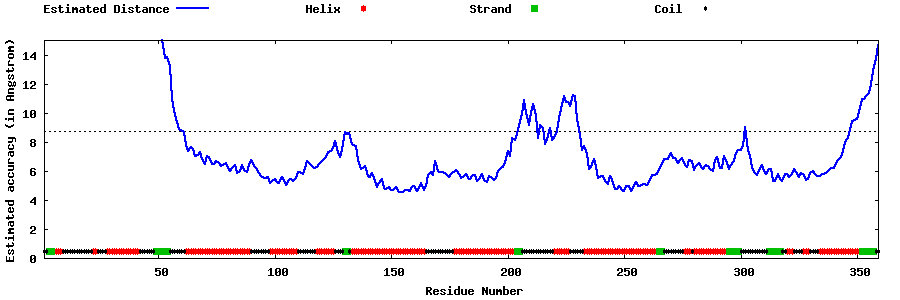 | |||
|
| |||
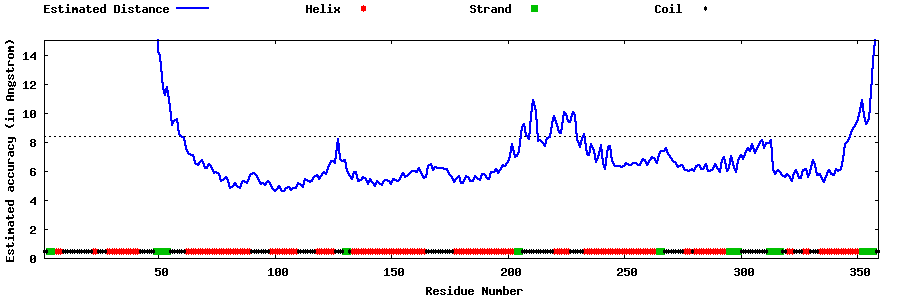 | |||
|
| |||
 | |||
|
| |||