

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MAQVRALHKIMALFSANSIGAMNNSDTRIAGCFLTGIPGLEQLHIWLSIPFCIMYITALEGNGILICVILSQAILHEPMYIFLSMLASADVLLSTTTMPKALANLWLGYSLISFDGCLTQMFFIHFLFIHSAVLLAMAFDRYVAICSPLRYVTILTSKVIGKIVTAALSHSFIIMFPSIFLLEHLHYCQINIIAHTFCEHMGIAHLSCSDISINVWYGLAAALLSTGLDIMLITVSYIHILQAVFRLLSQDARSKALSTCGSHICVILLFYVPALFSVFAYRFGGRSVPCYVHILLASLYVVIPPMLNPVIYGVRTKPILEGAKQMFSNLAKGSK | |
| CCHHHHHHHHHHHCCHHHCCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHHCCCCCC | |
| 92467778898863473334768998825324057998986688999999999999999999998998847887543189999999999998998559999999956998688878999999999988999999999812055115665450318889999999999999999998899999518999999448712112767888247836768999999999999999999999999999981788875888986505999999999999899998552268888984689999999973786646812215479999999999853344689 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MAQVRALHKIMALFSANSIGAMNNSDTRIAGCFLTGIPGLEQLHIWLSIPFCIMYITALEGNGILICVILSQAILHEPMYIFLSMLASADVLLSTTTMPKALANLWLGYSLISFDGCLTQMFFIHFLFIHSAVLLAMAFDRYVAICSPLRYVTILTSKVIGKIVTAALSHSFIIMFPSIFLLEHLHYCQINIIAHTFCEHMGIAHLSCSDISINVWYGLAAALLSTGLDIMLITVSYIHILQAVFRLLSQDARSKALSTCGSHICVILLFYVPALFSVFAYRFGGRSVPCYVHILLASLYVVIPPMLNPVIYGVRTKPILEGAKQMFSNLAKGSK | |
| 65423313301302333303321324231210200000121320200011012012202321210000020141001000200010010000002000020000000203403040000001101102200100010020000000200100000023100100000022120112021000210310452000000010210010002303001110020033133322310320111002000100146013201200100100000032213200100002433002000000022100300330030000203400410030025346648 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHHHHHHHHHCCHHHCCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHHCCCCCC MAQVRALHKIMALFSANSIGAMNNSDTRIAGCFLTGIPGLEQLHIWLSIPFCIMYITALEGNGILICVILSQAILHEPMYIFLSMLASADVLLSTTTMPKALANLWLGYSLISFDGCLTQMFFIHFLFIHSAVLLAMAFDRYVAICSPLRYVTILTSKVIGKIVTAALSHSFIIMFPSIFLLEHLHYCQINIIAHTFCEHMGIAHLSCSDISINVWYGLAAALLSTGLDIMLITVSYIHILQAVFRLLSQDARSKALSTCGSHICVILLFYVPALFSVFAYRFGGRSVPCYVHILLASLYVVIPPMLNPVIYGVRTKPILEGAKQMFSNLAKGSK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.22 | 0.83 | 2.26 | Download | ---------------------------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAAHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.25 | 0.80 | 2.03 | Download | -----------------------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 3 | 5tgzA | 0.17 | 0.25 | 0.84 | 1.81 | Download | -----------------------GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID-----------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 4 | 4ib4 | 0.17 | 0.22 | 0.84 | 1.51 | Download | -------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIE-T-----NPNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAAISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--- | |||||||||||||||||||
| 5 | 3uonA | 0.20 | 0.20 | 0.81 | 1.13 | Download | ----------------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQF--IVGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKS----RREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPN-TVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.22 | 0.82 | 2.52 | Download | -----------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 7 | 4iaq | 0.17 | 0.22 | 0.78 | 1.70 | Download | -----------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVNT--------------------DHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH----L-AIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 8 | 2z73A | 0.18 | 0.18 | 0.90 | 3.18 | Download | -------------------ETWWYNPSIVVHPHWREFDQVDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWIGFAACKVYGFIGGFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFG-----WGAEGVLCNFDY---------ISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQ | |||||||||||||||||||
| 9 | 5tgzA | 0.17 | 0.25 | 0.85 | 2.86 | Download | -----------------GGRGENFMDIEC--FMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 10 | 4gpoA | 0.21 | 0.18 | 0.83 | 4.10 | Download | ---------------------------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP-DWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL--------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
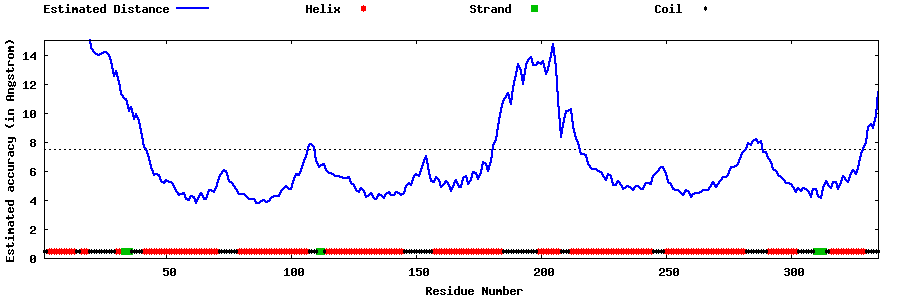
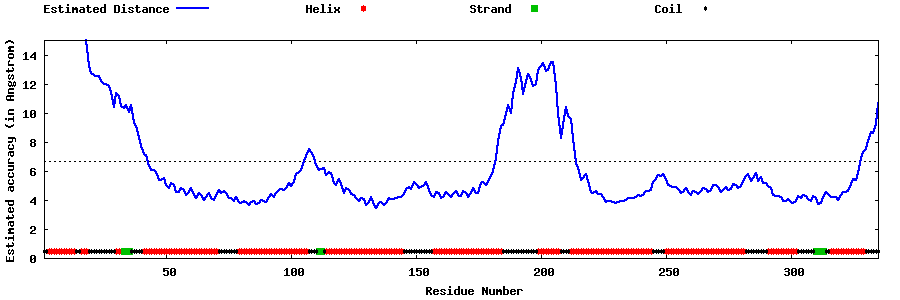
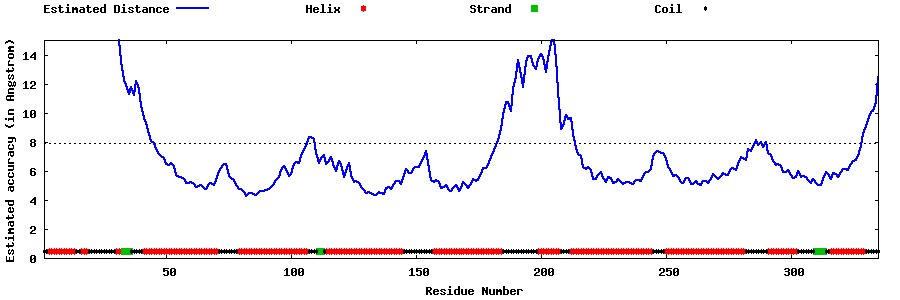
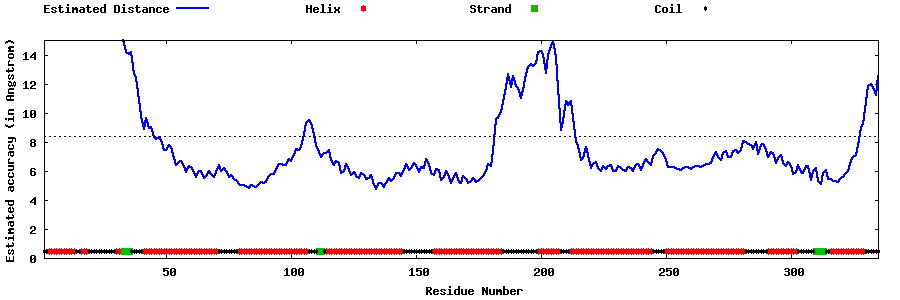
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||