

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDVGNKSTMSEFVLLGLSNSWELQMFFFMVFSLLYVATMVGNSLIVITVIVDPHLHSPMYFLLTNLSIIDMSLASFATPKMITDYLTGHKTISFDGCLTQIFFLHLFTGTEIILLMAMSFDRYIAICKPLHYASVISPQVCVALVVASWIMGVMHSMSQVIFALTLPFCGPYEVDSFFCDLPVVFQLACVDTYVLGLFMISTSGIIALSCFIVLFNSYVIVLVTVKHHSSRGSSKALSTCTAHFIVVFLFFGPCIFIYMWPLSSFLTDKILSVFYTIFTPTLNPIIYTLRNQEVKIAMRKLKNRFLNFNKAMPS | |
| CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCCCSSHHHHHHHHHHCCCHHHCCCCHHHHHHHHHHHHHHCCCCCCCCC | |
| 99877871357777359998315899999999999999987887444122179999978999873998867533346199999873599678589999999999999999999999998650787362203612267759999999999999999999999934589989884778632808888884034045463476665699999999999999999999730273123099999888999322663063258878899997521004577523440104743356299999999999852875687898 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDVGNKSTMSEFVLLGLSNSWELQMFFFMVFSLLYVATMVGNSLIVITVIVDPHLHSPMYFLLTNLSIIDMSLASFATPKMITDYLTGHKTISFDGCLTQIFFLHLFTGTEIILLMAMSFDRYIAICKPLHYASVISPQVCVALVVASWIMGVMHSMSQVIFALTLPFCGPYEVDSFFCDLPVVFQLACVDTYVLGLFMISTSGIIALSCFIVLFNSYVIVLVTVKHHSSRGSSKALSTCTAHFIVVFLFFGPCIFIYMWPLSSFLTDKILSVFYTIFTPTLNPIIYTLRNQEVKIAMRKLKNRFLNFNKAMPS | |
| 86464423000000000042440010001313332332333233000001004303000000023001200110000002000000253210005000000011033113102000200310000002211010000331000001201310220130013102303000012200000022100300002023000300230131023103313311210020013214612220010030021003023210100000033323301100030023103311300001054023003301432033456368 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCCCSSHHHHHHHHHHCCCHHHCCCCHHHHHHHHHHHHHHCCCCCCCCC MDVGNKSTMSEFVLLGLSNSWELQMFFFMVFSLLYVATMVGNSLIVITVIVDPHLHSPMYFLLTNLSIIDMSLASFATPKMITDYLTGHKTISFDGCLTQIFFLHLFTGTEIILLMAMSFDRYIAICKPLHYASVISPQVCVALVVASWIMGVMHSMSQVIFALTLPFCGPYEVDSFFCDLPVVFQLACVDTYVLGLFMISTSGIIALSCFIVLFNSYVIVLVTVKHHSSRGSSKALSTCTAHFIVVFLFFGPCIFIYMWPLSSFLTDKILSVFYTIFTPTLNPIIYTLRNQEVKIAMRKLKNRFLNFNKAMPS | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.21 | 0.87 | 3.30 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYFN-----------FFACVLVPLLLMLGVYLRIFLAARQLVHAAKSLAIIVGLFALCWLPLHIIN-CFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----- | |||||||||||||||||||
| 2 | 5tgzA | 0.18 | 0.24 | 0.89 | 2.23 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL---------QSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.20 | 0.23 | 0.80 | 2.09 | Download | ------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISILYTVYSTVGAF---------------------------------------YFPTLLLIALYGRIYVEARSRIIQMAARERKATKTLGIILGAFIVCWLPFFIISLVMPLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--------- | |||||||||||||||||||
| 4 | 4djh | 0.19 | 0.24 | 0.89 | 1.54 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSLSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.23 | 0.90 | 1.22 | Download | LKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------------ | |||||||||||||||||||
| 6 | 5tgzA | 0.18 | 0.24 | 0.89 | 3.46 | Download | GRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV--------LPLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------ | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.22 | 0.83 | 1.72 | Download | ---------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASE----------CVVNT-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADRERKATKTLGIILGAFIVCWLPFFIISLVMPIHL-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF---------- | |||||||||||||||||||
| 8 | 2z73A | 0.13 | 0.17 | 0.97 | 2.81 | Download | WY--NPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGTLEGVLCN------CSFDTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAK-RLNAKERLAKISIVIVSQFLLSWSPYAVVALLAQFGPWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDK | |||||||||||||||||||
| 9 | 5tgzA | 0.17 | 0.24 | 0.89 | 4.59 | Download | GRGENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-FAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------ | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.19 | 0.92 | 5.45 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFK | |||||||||||||||||||
| ||||||||||||||||||||||||||
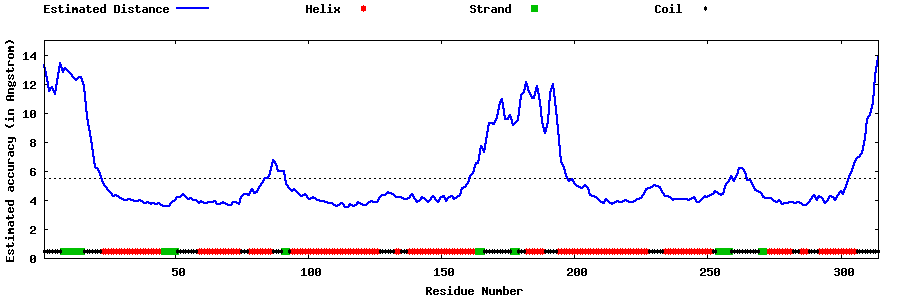
| Generated 3D models | Estimated local accuracy of models | ||
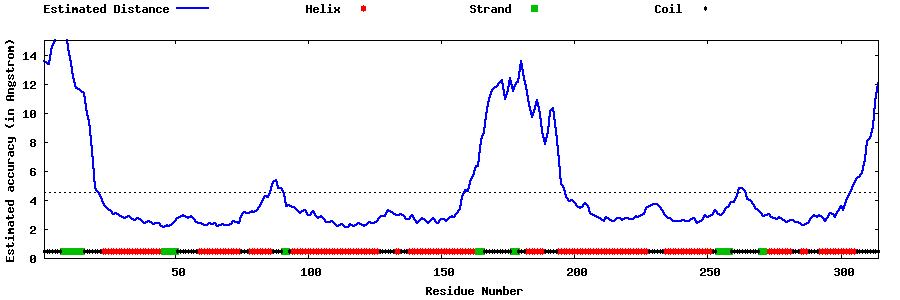 | |||
|
|
|||
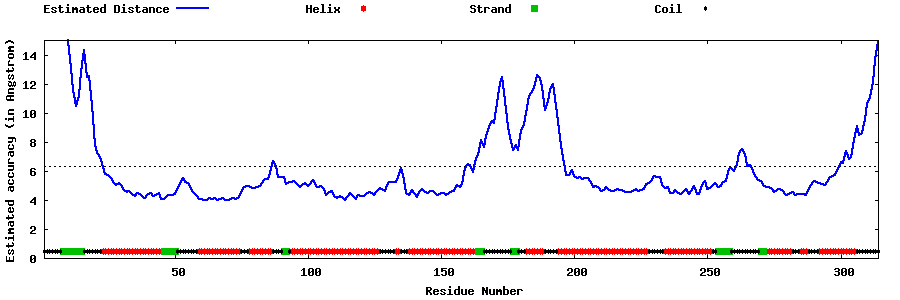 | |||
|
| |||
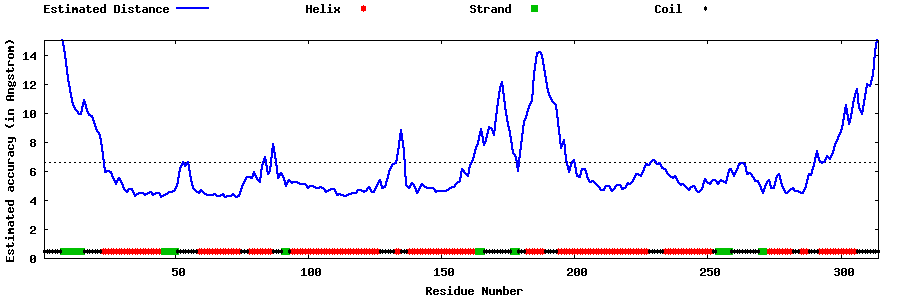 | |||
|
| |||
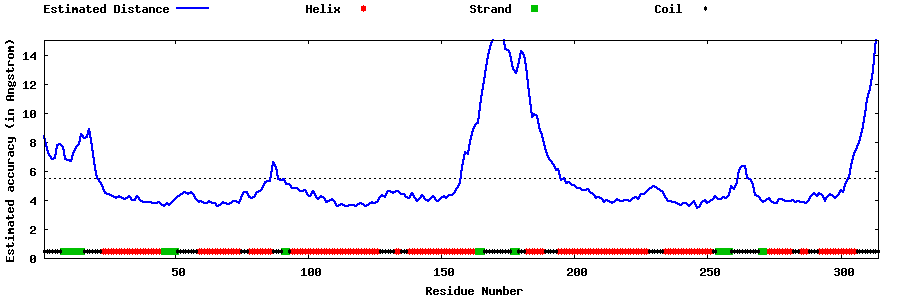 | |||
|
| |||
 | |||
|
| |||