

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDSLNQTRVTEFVFLGLTDNRVLEMLFFMAFSAIYMLTLSGNILIIIATVFTPSLHTPMYFFLSNLSFIDICHSSVTVPKMLEGLLLERKTISFDNCITQLFFLHLFACAEIFLLIIVAYDRYVAICTPLHYPNVMNMRVCIQLVFALWLGGTVHSLGQTFLTIRLPYCGPNIIDSYFCDVPLVIKLACTDTYLTGILIVTNSGTISLSCFLAVVTSYMVILVSLRKHSAEGRQKALSTCSAHFMVVALFFGPCIFIYTRPDTSFSIDKVVSVFYTVVTPLLNPFIYTLRNEEVKSAMKQLRQRQVFFTKSYT | |
| CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCHHHHHHHHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCCCC | |
| 9987787115777825999831689999999999999999788745534307999997899987599987623245738999987349967868999999999999999999999999866178636210262216775999999999999999999999993318988988467863283898888412314445447766669999999999999999999951037302219998988899916577315103884889999632433688762244010374335619999999999972357445779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDSLNQTRVTEFVFLGLTDNRVLEMLFFMAFSAIYMLTLSGNILIIIATVFTPSLHTPMYFFLSNLSFIDICHSSVTVPKMLEGLLLERKTISFDNCITQLFFLHLFACAEIFLLIIVAYDRYVAICTPLHYPNVMNMRVCIQLVFALWLGGTVHSLGQTFLTIRLPYCGPNIIDSYFCDVPLVIKLACTDTYLTGILIVTNSGTISLSCFLAVVTSYMVILVSLRKHSAEGRQKALSTCSAHFMVVALFFGPCIFIYTRPDTSFSIDKVVSVFYTVVTPLLNPFIYTLRNEEVKSAMKQLRQRQVFFTKSYT | |
| 8566342200000000004245001000131333233333323300000200430400001002300120011000000200000025321000500000001103311310200020030000000221101000023100000120131022013001310230300001220000002210030000202300030022013102310331331121002001321461222001003002100302321010000003331330100003102310331030000205402300330133203246628 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCHHHHHHHHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCCCC MDSLNQTRVTEFVFLGLTDNRVLEMLFFMAFSAIYMLTLSGNILIIIATVFTPSLHTPMYFFLSNLSFIDICHSSVTVPKMLEGLLLERKTISFDNCITQLFFLHLFACAEIFLLIIVAYDRYVAICTPLHYPNVMNMRVCIQLVFALWLGGTVHSLGQTFLTIRLPYCGPNIIDSYFCDVPLVIKLACTDTYLTGILIVTNSGTISLSCFLAVVTSYMVILVSLRKHSAEGRQKALSTCSAHFMVVALFFGPCIFIYTRPDTSFSIDKVVSVFYTVVTPLLNPFIYTLRNEEVKSAMKQLRQRQVFFTKSYT | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.21 | 0.88 | 3.36 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYF-----------NFFACVLVPLLLMLGVYLRIFLAARQLVHAAKSLAIIVGLFALCWLPLHIIN-CFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ---- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.21 | 0.89 | 2.28 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL---------QSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM------------ | |||||||||||||||||||
| 3 | 5tgzA | 0.16 | 0.21 | 0.86 | 1.91 | Download | GRGENFMD---------IENPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVCSDIF-----------------------PHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSMDIELAKTLVLILVVLIICWGPLLAIMVGKMNLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------- | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.22 | 0.89 | 1.53 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSLSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------- | |||||||||||||||||||
| 5 | 4yay | 0.20 | 0.25 | 0.90 | 1.23 | Download | LKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----------- | |||||||||||||||||||
| 6 | 5tgzA | 0.19 | 0.21 | 0.90 | 3.49 | Download | GRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV--------LPLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------- | |||||||||||||||||||
| 7 | 4iaq | 0.22 | 0.23 | 0.84 | 1.75 | Download | ---------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADRERKATKTLGIILGAFIVCWLPFFIISLVMPIHL-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-------- | |||||||||||||||||||
| 8 | 4ea3A | 0.18 | 0.19 | 0.83 | 2.96 | Download | -------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAIMGSAQV----------EDEEIECLVEIP------TPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGSAVFVGCWTPVQVFVLAQGLG----VQPSSTAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR-------------- | |||||||||||||||||||
| 9 | 5tgzA | 0.19 | 0.21 | 0.89 | 4.65 | Download | GRGENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVF-HRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------- | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.22 | 0.92 | 5.47 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPF | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
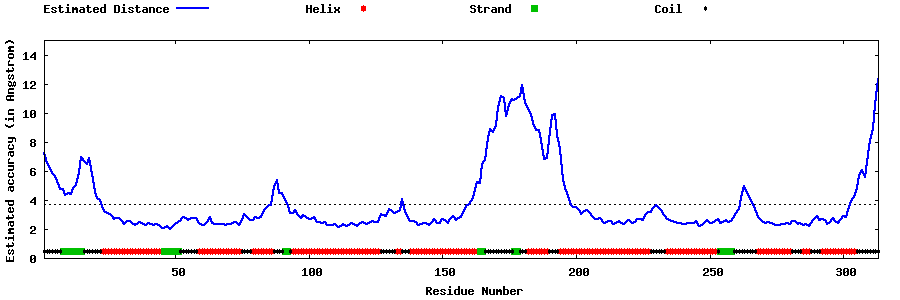 | |||
|
|
|||
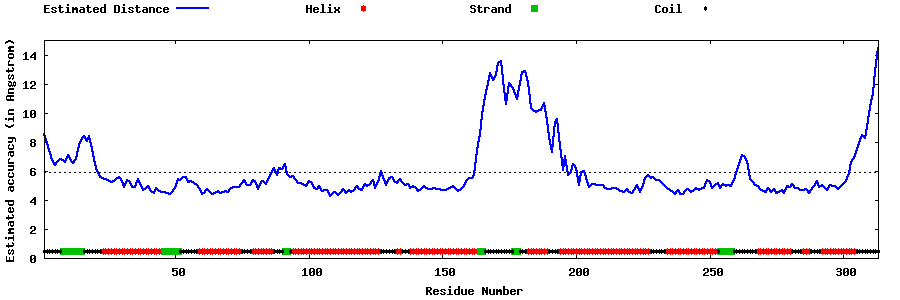 | |||
|
| |||
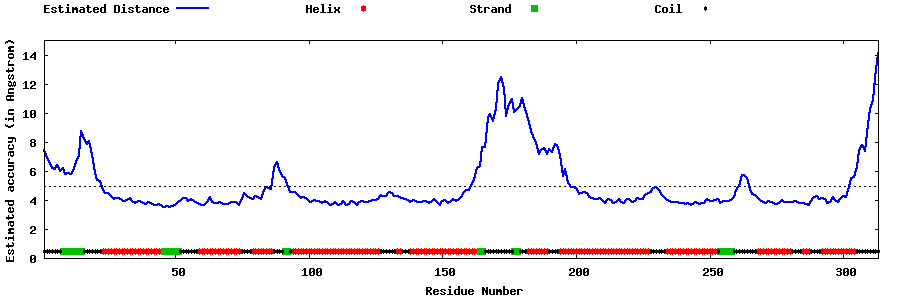 | |||
|
| |||
 | |||
|
| |||
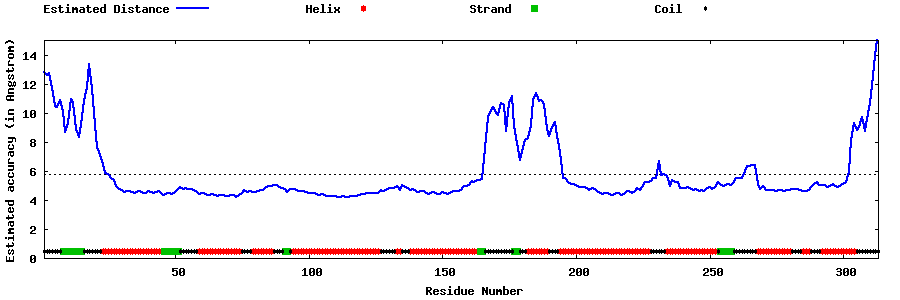 | |||
|
| |||