

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MTEFHLQSQMPSIRLIFRRLSLGRIKPSQSPRCSTSFMVVPSFSIAEHWRRMKGANLSQGMEFELLGLTTDPQLQRLLFVVFLGMYTATLLGNLVMFLLIHVSATLHTPMYSLLKSLSFLDFCYSSTVVPQTLVNFLAKRKVISYFGCMTQMFFYAGFATSECYLIAAMAYDRYAAICNPLLYSTIMSPEVCASLIVGSYSAGFLNSLIHTGCIFSLKFCGAHVVTHFFCDGPPILSLSCVDTSLCEILLFIFAGFNLLSCTLTILISYFLILNTILKMSSAQGRFKAFSTCASHLTAICLFFGTTLFMYLRPRSSYSLTQDRTVAVIYTVVIPVLNPLMYSLRNKDVKKALIKVWGRKTME | |
| CCCSSSHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHCCCCCCCCHHHHHHHCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCCHHCCCCHHHCCCCHHHHHHHHHHHHHCCCC | |
| 96131013315789999984235477655865310110133346524677511578842203468647999253899999999999999999989999996288766749888777998844721257199999871489788689999999999999999999999998651452062101880167889999999999999999999999984640889990487415808888762568627788899999999999899999999999999812677677654300548889979998732314582789988787897899882210312435644046598999999998520088 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MTEFHLQSQMPSIRLIFRRLSLGRIKPSQSPRCSTSFMVVPSFSIAEHWRRMKGANLSQGMEFELLGLTTDPQLQRLLFVVFLGMYTATLLGNLVMFLLIHVSATLHTPMYSLLKSLSFLDFCYSSTVVPQTLVNFLAKRKVISYFGCMTQMFFYAGFATSECYLIAAMAYDRYAAICNPLLYSTIMSPEVCASLIVGSYSAGFLNSLIHTGCIFSLKFCGAHVVTHFFCDGPPILSLSCVDTSLCEILLFIFAGFNLLSCTLTILISYFLILNTILKMSSAQGRFKAFSTCASHLTAICLFFGTTLFMYLRPRSSYSLTQDRTVAVIYTVVIPVLNPLMYSLRNKDVKKALIKVWGRKTME | |
| 64312133313303200342334314435444333322213313244314402650302001000000043240101001303321321232231001001000300000000010000000100000002000200154230103000000100000000000000000000000001001000300230000001000010010010000000202014322000000012000200012130000000100131133023003302210010001030262231000000002100001310110000003243124341000011032002300300002133014002201543428 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCSSSHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHCCCCCCCCHHHHHHHCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCCHHCCCCHHHCCCCHHHHHHHHHHHHHCCCC MTEFHLQSQMPSIRLIFRRLSLGRIKPSQSPRCSTSFMVVPSFSIAEHWRRMKGANLSQGMEFELLGLTTDPQLQRLLFVVFLGMYTATLLGNLVMFLLIHVSATLHTPMYSLLKSLSFLDFCYSSTVVPQTLVNFLAKRKVISYFGCMTQMFFYAGFATSECYLIAAMAYDRYAAICNPLLYSTIMSPEVCASLIVGSYSAGFLNSLIHTGCIFSLKFCGAHVVTHFFCDGPPILSLSCVDTSLCEILLFIFAGFNLLSCTLTILISYFLILNTILKMSSAQGRFKAFSTCASHLTAICLFFGTTLFMYLRPRSSYSLTQDRTVAVIYTVVIPVLNPLMYSLRNKDVKKALIKVWGRKTME | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.17 | 0.76 | 3.08 | Download | -----------------------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.20 | 0.76 | 1.83 | Download | ----------------------------------------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------- | |||||||||||||||||||
| 3 | 4iaqA | 0.19 | 0.18 | 0.74 | 1.85 | Download | ---------------------------------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVV------------------NTDHILYTVYSTVGAF--YFPTLLLIALYGRIYVEARSRIIQLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 4 | 4djh | 0.12 | 0.21 | 0.77 | 1.53 | Download | ----------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPD----DDSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---- | |||||||||||||||||||
| 5 | 4yay | 0.15 | 0.22 | 0.93 | 1.27 | Download | LNDNLKVIEKADVKDALTKMRQKATPPKLSPEMFRHGFLANEGKVKEAQEQLKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIH-RNVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------ | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.17 | 0.76 | 3.37 | Download | ------------------------------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 7 | 4iaq | 0.21 | 0.19 | 0.73 | 1.72 | Download | ------------------------------------------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 8 | 2z73A | 0.17 | 0.21 | 0.82 | 2.81 | Download | ----------------------------------ETWWYNPSIVVHPHWRE----------------FDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVL----------------CSRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTC | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.17 | 0.76 | 4.43 | Download | -----------------------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.15 | 0.79 | 5.05 | Download | --------------------------------------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| ||||||||||||||||||||||||||
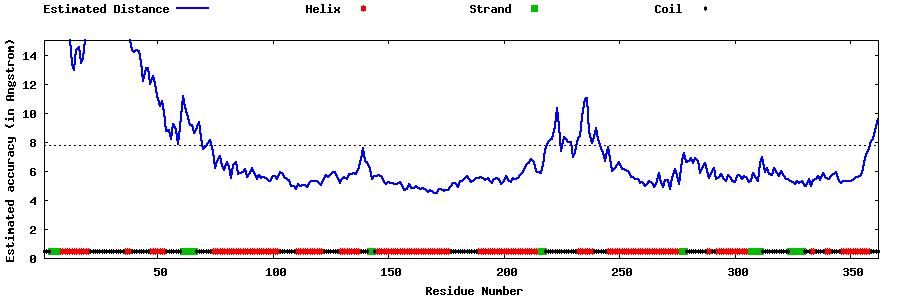
| Generated 3D models | Estimated local accuracy of models | ||
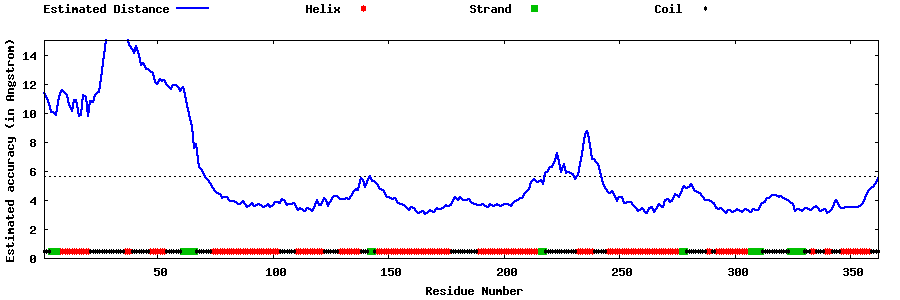 | |||
|
|
|||
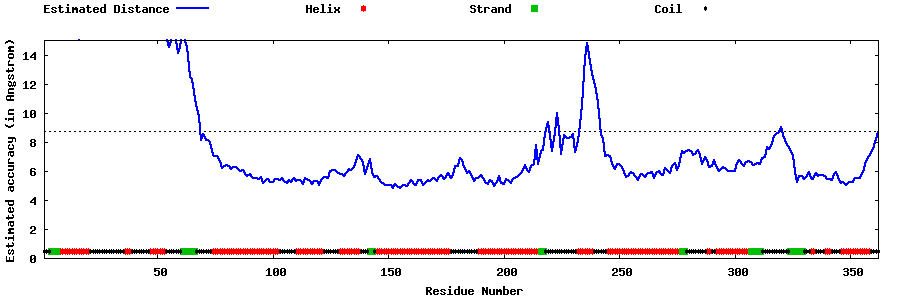 | |||
|
| |||
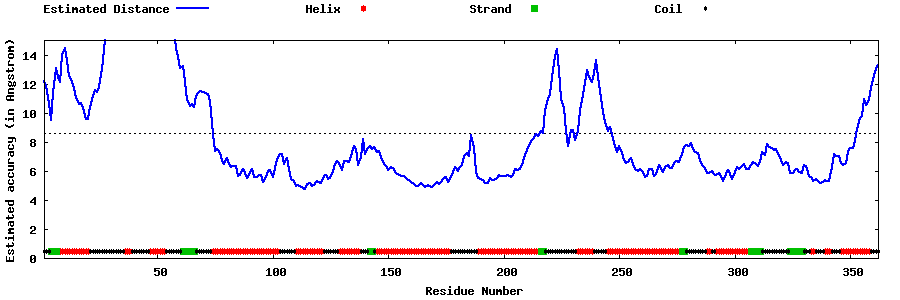 | |||
|
| |||
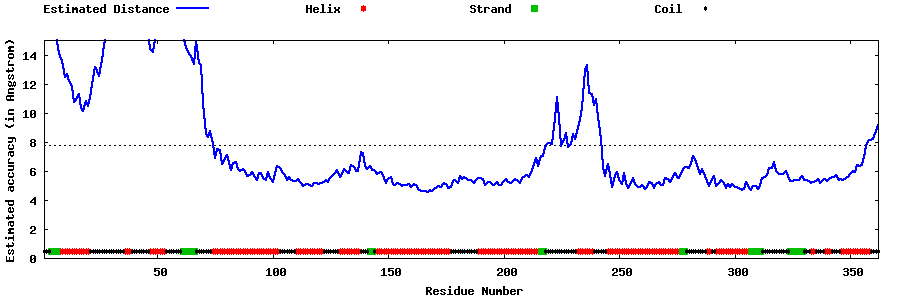 | |||
|
| |||
 | |||
|
| |||