

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDEANHSVVSEFVFLGLSDSRKIQLLLFLFFSVFYVSSLMGNLLIVLTVTSDPRLQSPMYFLLANLSIINLVFCSSTAPKMIYDLFRKHKTISFGGCVVQIFFIHAVGGTEMVLLIAMAFDRYVAICKPLHYLTIMNPQRCILFLVISWIIGIIHSVIQLAFVVDLLFCGPNELDSFFCDLPRFIKLACIETYTLGFMVTANSGFISLASFLILIISYIFILVTVQKKSSGGIFKAFSMLSAHVIVVVLVFGPLIFFYIFPFPTSHLDKFLAIFDAVITPVLNPVIYTFRNKEMMVAMRRRCSQFVNYSKIF | |
| CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCHHHCC | |
| 998778720588896799883258999999999999999877872563541799999789998859998787542366999998547996685899999999999999999999999986618874733146230687499999999999999999999999620899998837677518188888940250577889999888999999999999999999998501631312999888879993526630632488688999975321367886266400018553662999999999998526740069 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDEANHSVVSEFVFLGLSDSRKIQLLLFLFFSVFYVSSLMGNLLIVLTVTSDPRLQSPMYFLLANLSIINLVFCSSTAPKMIYDLFRKHKTISFGGCVVQIFFIHAVGGTEMVLLIAMAFDRYVAICKPLHYLTIMNPQRCILFLVISWIIGIIHSVIQLAFVVDLLFCGPNELDSFFCDLPRFIKLACIETYTLGFMVTANSGFISLASFLILIISYIFILVTVQKKSSGGIFKAFSMLSAHVIVVVLVFGPLIFFYIFPFPTSHLDKFLAIFDAVITPVLNPVIYTFRNKEMMVAMRRRCSQFVNYSKIF | |
| 866624230000000000424400100013133323323332330000020043030000000230012002100000020000002532100050000000110331131020002003100000022110100013310000012013102301300131023030000122000100221003000020230103002201310231033033112100200132146122200100300210030132101000000333233011100310231033113000121540230033014330324523 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCHHHCC MDEANHSVVSEFVFLGLSDSRKIQLLLFLFFSVFYVSSLMGNLLIVLTVTSDPRLQSPMYFLLANLSIINLVFCSSTAPKMIYDLFRKHKTISFGGCVVQIFFIHAVGGTEMVLLIAMAFDRYVAICKPLHYLTIMNPQRCILFLVISWIIGIIHSVIQLAFVVDLLFCGPNELDSFFCDLPRFIKLACIETYTLGFMVTANSGFISLASFLILIISYIFILVTVQKKSSGGIFKAFSMLSAHVIVVVLVFGPLIFFYIFPFPTSHLDKFLAIFDAVITPVLNPVIYTFRNKEMMVAMRRRCSQFVNYSKIF | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.22 | 0.88 | 3.33 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYF-----------NFFACVLVPLLLMLGVYLRIFLAARRQVHAAKSLAIIVGLFALCWLPLH-IINCFTFFCPDCAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 2 | 5tgzA | 0.18 | 0.23 | 0.89 | 2.25 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL---------QSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM----------- | |||||||||||||||||||
| 3 | 4iaqA | 0.19 | 0.20 | 0.83 | 2.17 | Download | ------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDHILYTVYSTVGAF--------------------YFPTLLLIALYGRIYVEARSLMAARERKATKTLGIILGAFIVCWLPFFIISLVM-PIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 4 | 4ib4 | 0.18 | 0.22 | 0.88 | 1.54 | Download | -----------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKG--IE--T--NPNNITCVLTKE---------RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----- | |||||||||||||||||||
| 5 | 4yay | 0.18 | 0.25 | 0.90 | 1.22 | Download | LKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------- | |||||||||||||||||||
| 6 | 5tgzA | 0.18 | 0.23 | 0.90 | 3.48 | Download | GRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV--------LPLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 7 | 4iaq | 0.17 | 0.21 | 0.84 | 1.73 | Download | --------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQAS----------ECVVNT-------DHILYTVYS---TVGAFYFPTLLLIALYGRIYVEARSRIADRERKATKTLGIILGAFIVCWLPFFIISLVMPIHL-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF-------- | |||||||||||||||||||
| 8 | 4buoA | 0.18 | 0.21 | 0.89 | 3.03 | Download | ----NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPI------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQAVVIAFVVCWLPYHVRRLMFCYI--------SDEQWTYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL---------- | |||||||||||||||||||
| 9 | 5tgzA | 0.18 | 0.23 | 0.89 | 4.65 | Download | GRGENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-FAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.23 | 0.92 | 5.43 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVAAENLY | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
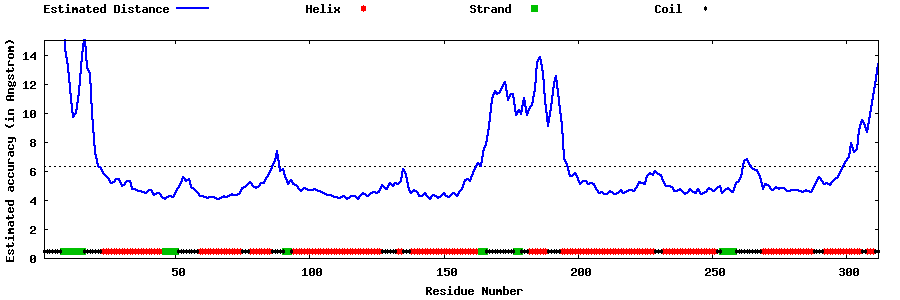 | |||
|
| |||
 | |||
|
| |||
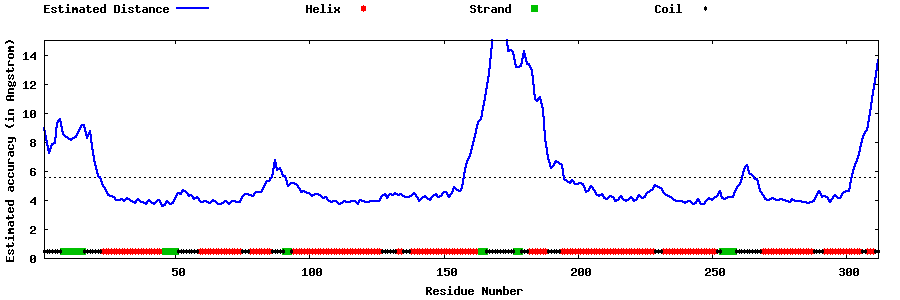 | |||
|
| |||
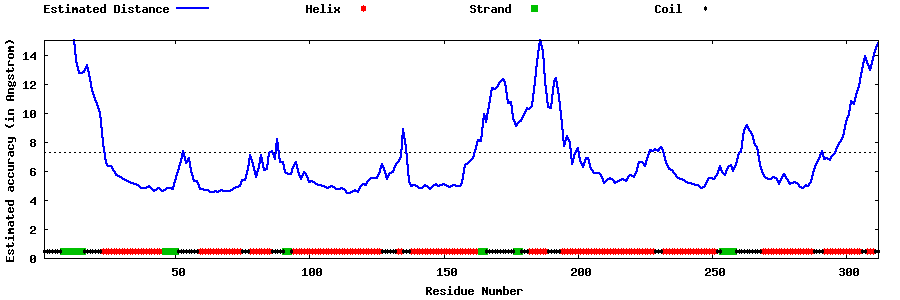 | |||
|
| |||