

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNISDVISFDILVSAMKTGNQSFGTDFLLVGLFQYGWINSLLFVVIATLFTVALTGNIMLIHLIRLNTRLHTPMYFLLSQLSIVDLMYISTTVPKMAVSFLSQSKTIRFLGCEIQTYVFLALGGTEALLLGFMSYDRYVAICHPLHYPMLMSKKICCLMVACAWASGSINAFIHTLYVFQLPFCRSRLINHFFCEVPALLSLVCQDTSQYEYTVLLSGLIILLLPFLAILASYARVLIVVFQMSSGKGQAKAVSTCSSHLIVASLFYATTLFTYTRPHSLRSPSRDKAVAVFYTIVTPLLNPFIYSLRNKEVTGAVRRLLGYWICCRKYDFRSLY | |
| CCHHHHHCHHHHHHHHHCCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCSSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHCCCCCCCCC | |
| 96445425698996611777820001268789998015889999999999999998999999998577745649999988999988776126899999834799578688899999999999999999999998650675165556884268869999999999999999999999994068899996888525848888897037419999999999999999899999999999999824876131542764468789989999875775252789999987782898327306543023543146499999999999751001114210259 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNISDVISFDILVSAMKTGNQSFGTDFLLVGLFQYGWINSLLFVVIATLFTVALTGNIMLIHLIRLNTRLHTPMYFLLSQLSIVDLMYISTTVPKMAVSFLSQSKTIRFLGCEIQTYVFLALGGTEALLLGFMSYDRYVAICHPLHYPMLMSKKICCLMVACAWASGSINAFIHTLYVFQLPFCRSRLINHFFCEVPALLSLVCQDTSQYEYTVLLSGLIILLLPFLAILASYARVLIVVFQMSSGKGQAKAVSTCSSHLIVASLFYATTLFTYTRPHSLRSPSRDKAVAVFYTIVTPLLNPFIYSLRNKEVTGAVRRLLGYWICCRKYDFRSLY | |
| 44132113122114303530201101000000043251210001203332331333322001001001300000011020000100111002002000010165310000002201110130031002000200200000002102000100330000001200200020030001100001003524010000022100300010023002102331233133113100201120010001031461231000000000000001320000000103143245331000011023213300300001042023002200232110442414425 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHHHCHHHHHHHHHCCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCSSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHCCCCCCCCC MNISDVISFDILVSAMKTGNQSFGTDFLLVGLFQYGWINSLLFVVIATLFTVALTGNIMLIHLIRLNTRLHTPMYFLLSQLSIVDLMYISTTVPKMAVSFLSQSKTIRFLGCEIQTYVFLALGGTEALLLGFMSYDRYVAICHPLHYPMLMSKKICCLMVACAWASGSINAFIHTLYVFQLPFCRSRLINHFFCEVPALLSLVCQDTSQYEYTVLLSGLIILLLPFLAILASYARVLIVVFQMSSGKGQAKAVSTCSSHLIVASLFYATTLFTYTRPHSLRSPSRDKAVAVFYTIVTPLLNPFIYSLRNKEVTGAVRRLLGYWICCRKYDFRSLY | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.18 | 0.83 | 3.32 | Download | -----------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-------- | |||||||||||||||||||
| 2 | 5tgzA | 0.18 | 0.22 | 0.84 | 2.18 | Download | ----------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL--------QSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM---------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.20 | 0.79 | 1.97 | Download | ---------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH-----------------ILYTVYSTVGAF---YFPTLLLIALYGRIYVEARSLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMP---IHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------ | |||||||||||||||||||
| 4 | 3uon | 0.18 | 0.24 | 0.82 | 1.56 | Download | -----------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFI--VGVRTVEDGECY----------IQFFNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIAREKKVTRTILAILLAFIITWAPYNVMVLNTFCPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.24 | 0.89 | 1.25 | Download | FDLANEGKVKEAQELKTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVF-F------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--------------- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.18 | 0.83 | 3.56 | Download | ------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-------- | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.20 | 0.77 | 1.72 | Download | ------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWLIISLVMPIH--LAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF------------- | |||||||||||||||||||
| 8 | 2z73A | 0.17 | 0.16 | 0.89 | 2.70 | Download | ----------------ETWNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGTLEGVLCNDYI----------------SRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQF-----D | |||||||||||||||||||
| 9 | 3emlA | 0.19 | 0.18 | 0.83 | 4.31 | Download | -----------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFT-FFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-------- | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.19 | 0.88 | 5.15 | Download | --------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFKAAA | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
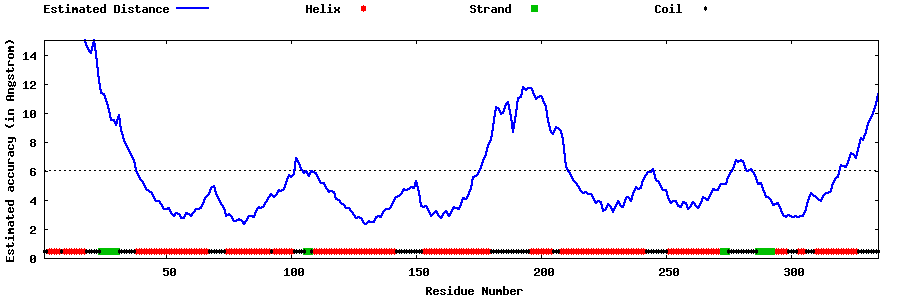 | |||
|
|
|||
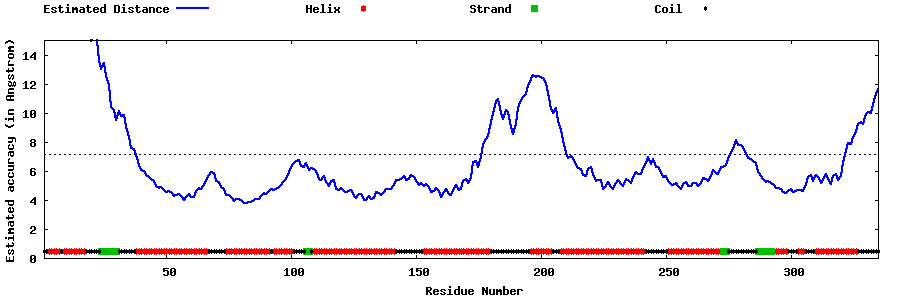 | |||
|
| |||
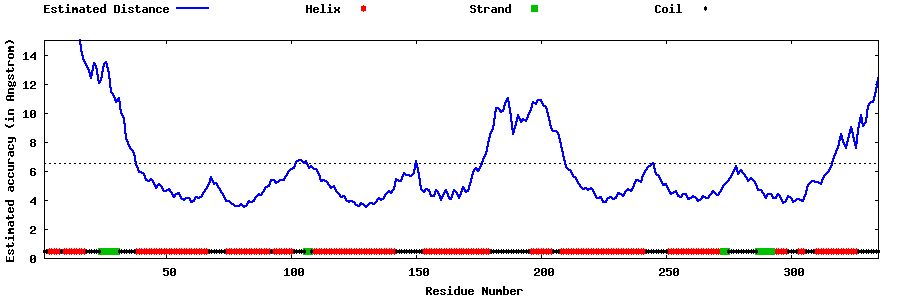 | |||
|
| |||
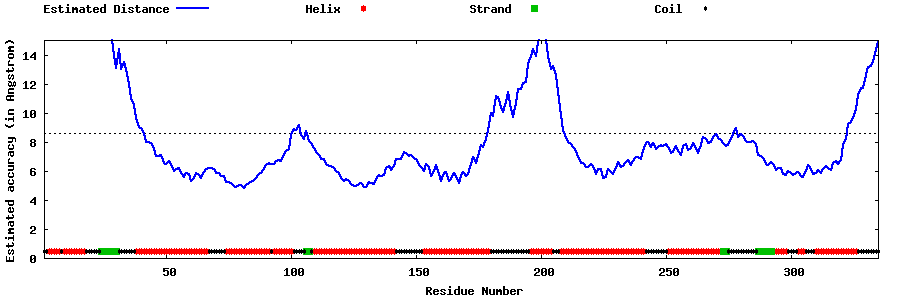 | |||
|
| |||
 | |||
|
| |||