

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MIIYKQGITFLQKENNNTIHLNTMFFLSPAETHQRMAAENHSFVTKFILVGLTEKSELQLPLFLVFLGIYVVTVLGNLGMITLIGLSSHLHTPMYCFLSSLSFIDFCHSTVITPKMLVNFVTEKNIISYPECMTQLYFFLVFAIAECHMLAAMAYDGYVAICSPLLYSIIISNKACFSLILVVYVIGLICASAHIGCMFRVQFCKFDVINHYFCDLISILKLSCSSTYINELLILIFSGINILVPSLTILSSYIFIIASILRIRYTEGRSKAFSTCSSHISAVSVFFGSAAFMYLQPSSVSSMDQGKVSSVFYTIVVPMLNPLIYSLRNKDVHVALKKTLGKRTFL | |
| CCSSHHCCCHHHHHCCCCCSSSSSSSCCCCCHHHHHHCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHCCCCC | |
| 9121001340345346530133455405774676520578831203578658899350899999999999999988879999985088877738888767998844721257088989872489788489999999999999999999999987541330162101880166789999999999999999999999984652789992385426818887670568636677888888999999999999999999999813666576765200648889979999744403484789998787897899881101412435644166498999999998422289 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MIIYKQGITFLQKENNNTIHLNTMFFLSPAETHQRMAAENHSFVTKFILVGLTEKSELQLPLFLVFLGIYVVTVLGNLGMITLIGLSSHLHTPMYCFLSSLSFIDFCHSTVITPKMLVNFVTEKNIISYPECMTQLYFFLVFAIAECHMLAAMAYDGYVAICSPLLYSIIISNKACFSLILVVYVIGLICASAHIGCMFRVQFCKFDVINHYFCDLISILKLSCSSTYINELLILIFSGINILVPSLTILSSYIFIIASILRIRYTEGRSKAFSTCSSHISAVSVFFGSAAFMYLQPSSVSSMDQGKVSSVFYTIVVPMLNPLIYSLRNKDVHVALKKTLGKRTFL | |
| 5101344142345434431313110212344334403550302001000000043260100001303331321232231002001000300000000020000000100000002000200154220003000000100000000000000000000000002111000200230000001101110020030100000202004421000000022000200013230101000110131133023003302310010001030261231010000002100001320110000003253224341000021032002300300002133014002200533316 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCSSHHCCCHHHHHCCCCCSSSSSSSCCCCCHHHHHHCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHCCCCC MIIYKQGITFLQKENNNTIHLNTMFFLSPAETHQRMAAENHSFVTKFILVGLTEKSELQLPLFLVFLGIYVVTVLGNLGMITLIGLSSHLHTPMYCFLSSLSFIDFCHSTVITPKMLVNFVTEKNIISYPECMTQLYFFLVFAIAECHMLAAMAYDGYVAICSPLLYSIIISNKACFSLILVVYVIGLICASAHIGCMFRVQFCKFDVINHYFCDLISILKLSCSSTYINELLILIFSGINILVPSLTILSSYIFIIASILRIRYTEGRSKAFSTCSSHISAVSVFFGSAAFMYLQPSSVSSMDQGKVSSVFYTIVVPMLNPLIYSLRNKDVHVALKKTLGKRTFL | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.19 | 0.80 | 3.25 | Download | -------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.18 | 0.79 | 1.80 | Download | ------------------------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------- | |||||||||||||||||||
| 3 | 4iaqA | 0.17 | 0.19 | 0.79 | 2.17 | Download | -----------------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPFFWR---------------ECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQKYAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 4 | 4djh | 0.13 | 0.17 | 0.81 | 1.54 | Download | ------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---- | |||||||||||||||||||
| 5 | 4yay | 0.15 | 0.21 | 0.92 | 1.28 | Download | ALDAQKDKSPDSPEMKDFRHGFDINEGKVKAAAEQLKTTRNAYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIH-RNVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------ | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.19 | 0.80 | 3.50 | Download | --------------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.17 | 0.76 | 1.72 | Download | --------------------------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVNT--------------------DHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIAAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 8 | 2z73A | 0.19 | 0.22 | 0.86 | 2.75 | Download | --------------------------------------ENPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDY---------ISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTC | |||||||||||||||||||
| 9 | 3emlA | 0.19 | 0.19 | 0.79 | 4.90 | Download | ---------------------------------------------------------ISSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.18 | 0.83 | 5.48 | Download | ----------------------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| ||||||||||||||||||||||||||
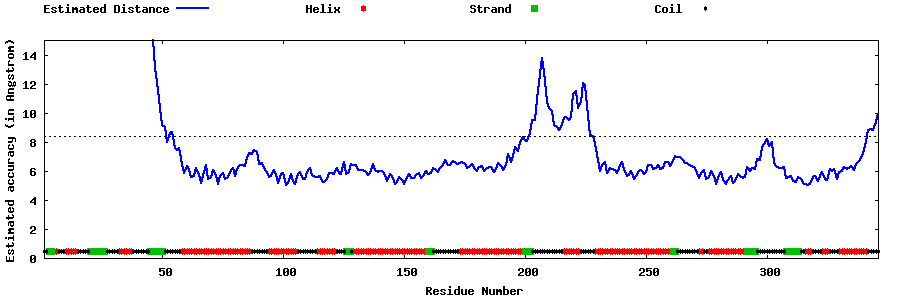
| Generated 3D models | Estimated local accuracy of models | ||
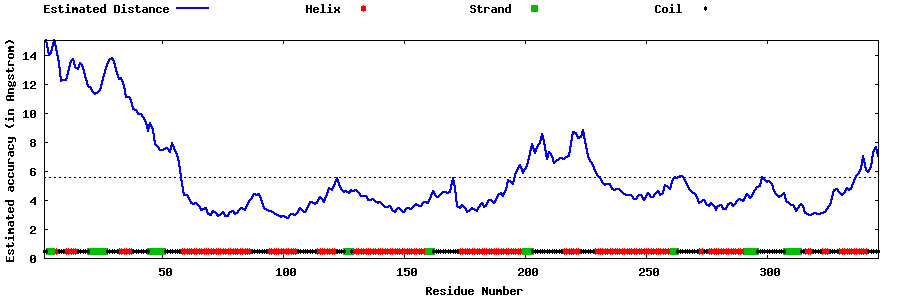 | |||
|
|
|||
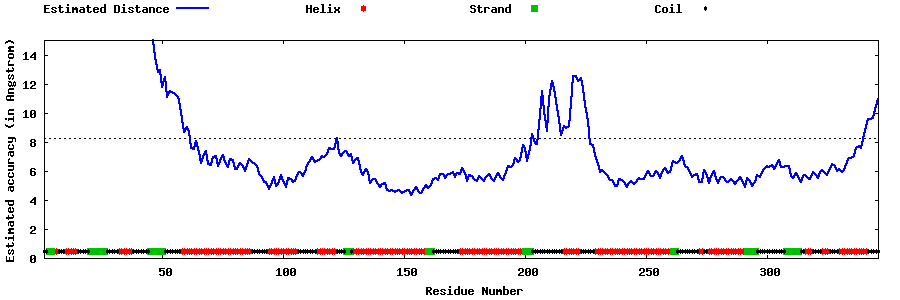 | |||
|
| |||
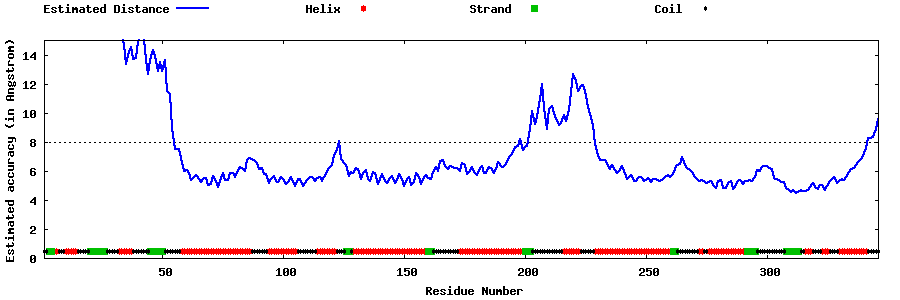 | |||
|
| |||
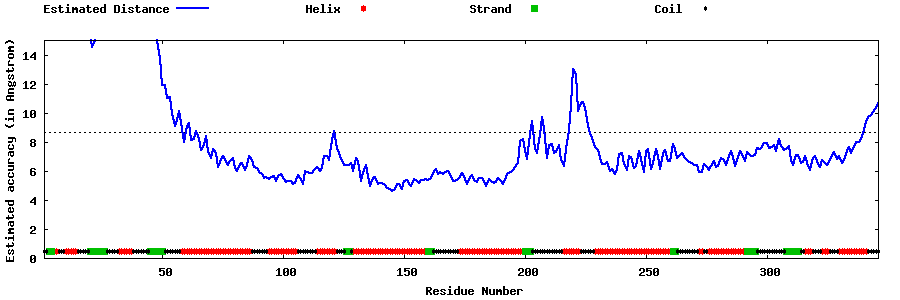 | |||
|
| |||
 | |||
|
| |||