

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MAAQNGNTSFTPNFNPPQDHASSLSFNFSYGDYDLPMDEDEDMTKTRTFFAAKIVIGIALAGIMLVCGIGNFVFIAALTRYKKLRNLTNLLIANLAISDFLVAIICCPFEMDYYVVRQLSWEHGHVLCASVNYLRTVSLYVSTNALLAIAIDRYLAIVHPLKPRMNYQTASFLIALVWMVSILIAIPSAYFATETVLFIVKSQEKIFCGQIWPVDQQLYYKSYFLFIFGVEFVGPVVTMTLCYARISRELWFKAVPGFQTEQIRKRLRCRRKTVLVLMCILTAYVLCWAPFYGFTIVRDFFPTVFVKEKHYLTAFYVVECIAMSNSMINTVCFVTVKNNTMKYFKKMMLLHWRPSQRGSKSSADLDLRTNGVPTTEEVDCIRLK | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 999999867899999877777777777787655678776544444465306899999999999999989999897988618998854126999999999999999718999999973798206558888899999999999999999999986999664487551151487720699999999999999991657866517998179984289713699999999999999999999999999999999815689876526678888852349999999999999997799999999997523255506999999999999999999999999980999999999997352478777888888878756998989988720169 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MAAQNGNTSFTPNFNPPQDHASSLSFNFSYGDYDLPMDEDEDMTKTRTFFAAKIVIGIALAGIMLVCGIGNFVFIAALTRYKKLRNLTNLLIANLAISDFLVAIICCPFEMDYYVVRQLSWEHGHVLCASVNYLRTVSLYVSTNALLAIAIDRYLAIVHPLKPRMNYQTASFLIALVWMVSILIAIPSAYFATETVLFIVKSQEKIFCGQIWPVDQQLYYKSYFLFIFGVEFVGPVVTMTLCYARISRELWFKAVPGFQTEQIRKRLRCRRKTVLVLMCILTAYVLCWAPFYGFTIVRDFFPTVFVKEKHYLTAFYVVECIAMSNSMINTVCFVTVKNNTMKYFKKMMLLHWRPSQRGSKSSADLDLRTNGVPTTEEVDCIRLK | |
| 754434321232313334332322222212332313343343133231220010000200120020023132000000000340210011002001302200010101010000013332000010001001000000000000000000000020000002332332100000000110011011000000202314224443111010201344420200000010221123101000200010001023342324446534433443200000000000000000223000000100234313432001001000000000000000001000042014002300201213444446434434343444434444334327 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MAAQNGNTSFTPNFNPPQDHASSLSFNFSYGDYDLPMDEDEDMTKTRTFFAAKIVIGIALAGIMLVCGIGNFVFIAALTRYKKLRNLTNLLIANLAISDFLVAIICCPFEMDYYVVRQLSWEHGHVLCASVNYLRTVSLYVSTNALLAIAIDRYLAIVHPLKPRMNYQTASFLIALVWMVSILIAIPSAYFATETVLFIVKSQEKIFCGQIWPVDQQLYYKSYFLFIFGVEFVGPVVTMTLCYARISRELWFKAVPGFQTEQIRKRLRCRRKTVLVLMCILTAYVLCWAPFYGFTIVRDFFPTVFVKEKHYLTAFYVVECIAMSNSMINTVCFVTVKNNTMKYFKKMMLLHWRPSQRGSKSSADLDLRTNGVPTTEEVDCIRLK | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.22 | 0.21 | 0.77 | 2.99 | Download | ----------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM----TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVD-IDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP--------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.22 | 0.20 | 0.78 | 3.82 | Download | ------------------------------------------------KETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAED--WPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDY-KGSYLRICLLHPVQKTAFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC---------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.21 | 0.78 | 3.57 | Download | ----------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMET--WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.18 | 0.22 | 0.91 | 1.52 | Download | ---------------DLRDNETWWYN-PSIIVH---PHWR---EFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNFPLMTISCFL--KKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYT----LEGVLCNCSFDYISRD-STTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEW----VTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKDAETEIPAGESSDAAPSADAAQMK | |||||||||||||||||||
| 5 | 4rnb | 0.28 | 0.23 | 0.76 | 1.15 | Download | ------------------------------------------------PKEYEWVLIAGYIIVFVVALIGNVLVCVAVWKNHHMRTVTNYFIVNLSLADVLVTITCLPATLVVDITE--TWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPST----AKRARNSIVIIWIVSCIIMIPQAIVMECSTVF--KTTLFTVCDERWGGE--IYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSLMSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMFAHDRETVYAWFTFSHWLVYANSAANPIIYNFLSGKFREEFKAAFSC---------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.22 | 0.21 | 0.78 | 3.24 | Download | ------------------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMET--WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLS----GSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI-DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.23 | 0.21 | 0.73 | 1.71 | Download | -----------------------------------------------------VFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTV--IGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVR--TVEDGECYIQFFSN-----AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGAWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIP-----NTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.22 | 0.20 | 0.78 | 4.75 | Download | ---------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMET--WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI-DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.21 | 0.21 | 0.78 | 3.17 | Download | ----------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM----TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVD-IDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.22 | 0.22 | 0.76 | 3.72 | Download | -------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLM--GTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR-----QGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS----KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALIT--IPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
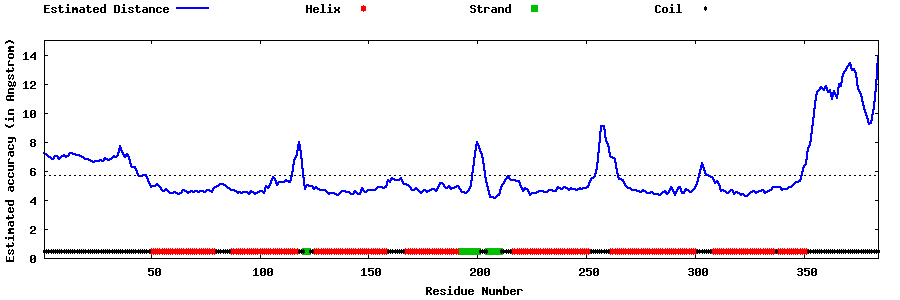
| Generated 3D models | Estimated local accuracy of models | ||
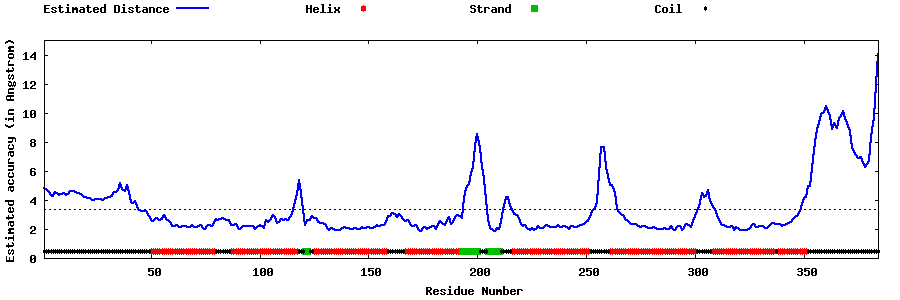 | |||
|
|
|||
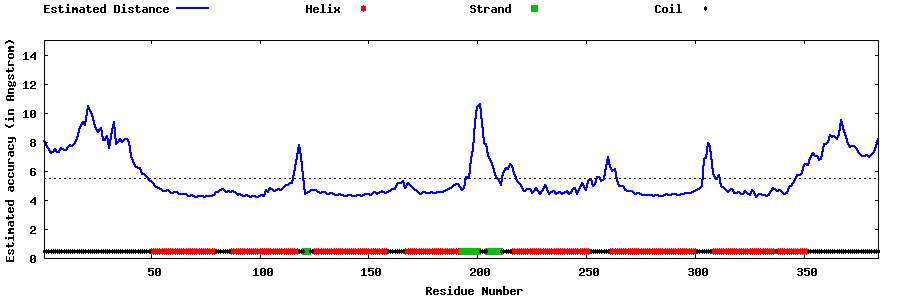 | |||
|
| |||
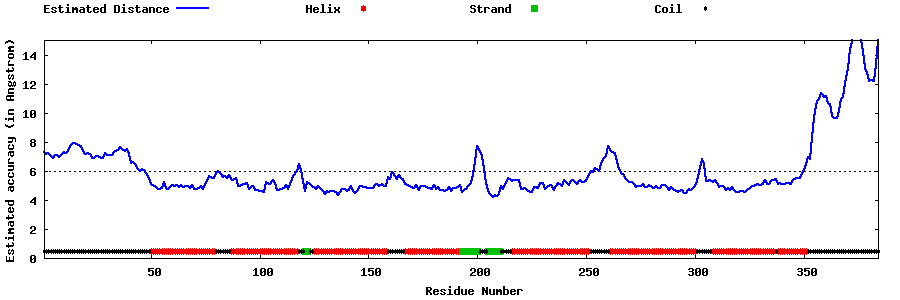 | |||
|
| |||
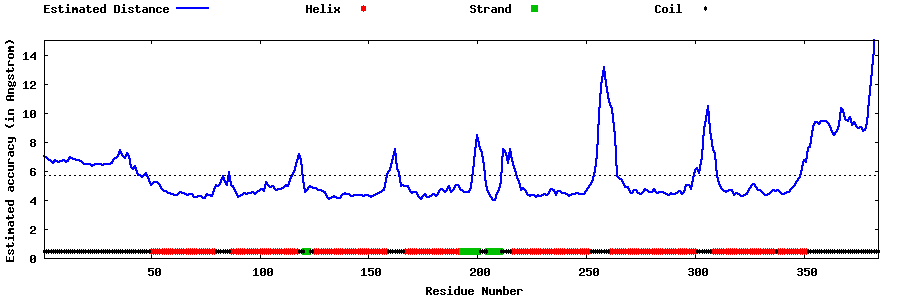 | |||
|
| |||
 | |||
|
| |||