

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MPGHNTSRNSSCDPIVTPHLISLYFIVLIGGLVGVISILFLLVKMNTRSVTTMAVINLVVVHSVFLLTVPFRLTYLIKKTWMFGLPFCKFVSAMLHIHMYLTFLFYVVILVTRYLIFFKCKDKVEFYRKLHAVAASAGMWTLVIVIVVPLVVSRYGIHEEYNEEHCFKFHKELAYTYVKIINYMIVIFVIAVAVILLVFQVFIIMLMVQKLRHSLLSHQEFWAQLKNLFFIGVILVCFLPYQFFRIYYLNVVTHSNACNSKVAFYNEIFLSVTAISCYDLLLFVFGGSHWFKQKIIGLWNCVLCR | |
| CCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHSSSSSSSHHHHHCCCHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCC | |
| 95999899988521257419999999999999999981043034488858899999999999999997999999983699885974687999999999999999999999999899742401133645024776778999999999999999730167799967981579813589999999998635599999999999999999871456653101032421247885511577605188999999985337886577999999999999999989999987068178899999997334259 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MPGHNTSRNSSCDPIVTPHLISLYFIVLIGGLVGVISILFLLVKMNTRSVTTMAVINLVVVHSVFLLTVPFRLTYLIKKTWMFGLPFCKFVSAMLHIHMYLTFLFYVVILVTRYLIFFKCKDKVEFYRKLHAVAASAGMWTLVIVIVVPLVVSRYGIHEEYNEEHCFKFHKELAYTYVKIINYMIVIFVIAVAVILLVFQVFIIMLMVQKLRHSLLSHQEFWAQLKNLFFIGVILVCFLPYQFFRIYYLNVVTHSNACNSKVAFYNEIFLSVTAISCYDLLLFVFGGSHWFKQKIIGLWNCVLCR | |
| 64743424513244011000022012003313321220010002234320000000000303201200010100000364010020001100121331111103100100320010001013034133321010000201110120010100022034574221101212464312101112111323333332012010100110133334563454442201000000010010012210200001011223354142111000000000031002100000000361124101400421238 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHSSSSSSSHHHHHCCCHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCC MPGHNTSRNSSCDPIVTPHLISLYFIVLIGGLVGVISILFLLVKMNTRSVTTMAVINLVVVHSVFLLTVPFRLTYLIKKTWMFGLPFCKFVSAMLHIHMYLTFLFYVVILVTRYLIFFKCKDKVEFYRKLHAVAASAGMWTLVIVIVVPLVVSRYGIHEEYNEEHCFKFHKELAYTYVKIINYMIVIFVIAVAVILLVFQVFIIMLMVQKLRHSLLSHQEFWAQLKNLFFIGVILVCFLPYQFFRIYYLNVVTHSNACNSKVAFYNEIFLSVTAISCYDLLLFVFGGSHWFKQKIIGLWNCVLCR | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.19 | 0.22 | 0.97 | 3.39 | Download | ----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK---PC | |||||||||||||||||||
| 2 | 4xnwA | 0.17 | 0.21 | 0.95 | 3.85 | Download | ---SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYLRFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKEEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQTNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.19 | 0.22 | 0.97 | 3.90 | Download | ----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-PLVVAALHLCIALGYANSSLNPVLYAFLD-ENFKRCFRQLCRKPCG- | |||||||||||||||||||
| 4 | 4djh | 0.18 | 0.23 | 0.90 | 1.53 | Download | ------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDLICVFIFAFVIPVLII------IVCYTLMILRLKSVRLLSEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA--------ALSSYYFCIALYTNSSLNPILYAFLDEN-FKRCFRDFCFP---- | |||||||||||||||||||
| 5 | 4xnv | 0.17 | 0.20 | 0.94 | 1.17 | Download | -----SFKCATKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHKPWSGISVYMFNLALADFLYVLTLPALIFYYFNTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYLYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYTVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDPAMCARVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.18 | 0.22 | 0.97 | 3.56 | Download | ------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-- | |||||||||||||||||||
| 7 | 4djh | 0.20 | 0.23 | 0.91 | 1.72 | Download | -------------PAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-------AALSSYYFCIALGYTNSSLNPILYAFLDEN-FKRCFRDFCF----- | |||||||||||||||||||
| 8 | 4mbsA | 0.17 | 0.20 | 0.93 | 3.76 | Download | -----PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTR-SQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK--------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------ | |||||||||||||||||||
| 9 | 5o9hA | 0.16 | 0.19 | 0.94 | 3.49 | Download | ----------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYPPKVLCGVDHD-KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARET------RSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKS-LPELLREVLTE | |||||||||||||||||||
| 10 | 5c1mA | 0.18 | 0.20 | 0.95 | 5.14 | Download | -GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQ-GSIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKERNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET--TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
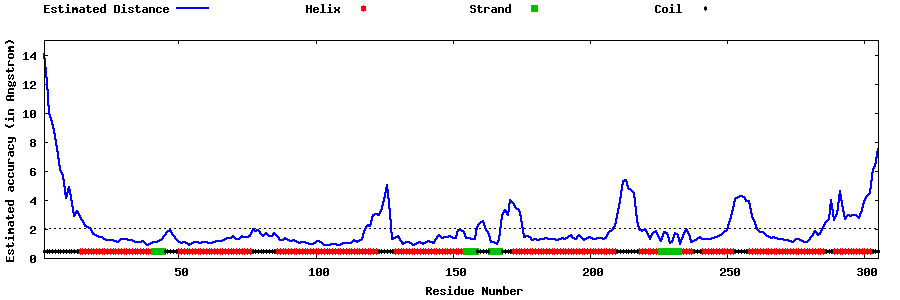 | |||
|
|
|||
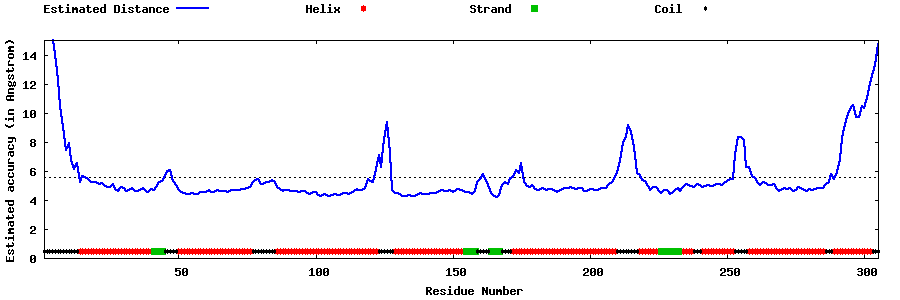 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||