

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MLKLVIIENMAEIMLFSLDLLLFSTDILCFNFPSKMIKLPGFITIQIFFYPQASFGISANTILLLFHIFTFVFSHRSKSIDMIISHLSLIHILLLFTQAILVSLDFFGSQNTQDDLRYKVIVFLNKVMRGLSICTPCLLSVLQAIISPSIFSLAKLKHPSASHILGFFLFSWVLNMFIGVIFCCTLRLPPVKRGQSSVCHTALFLFAHELHPQETVFHTNDFEGCHLYRVHGPLKRLHGDYFIQTIRGYLSAFTQPACPRVSPVKRASQAILLLVSFVFTYWVDFTFSFSGGVTWINDSLLVWLQVIVANSYAAISPLMLIYADNQIFKTLQMLWFKYLSPPKLMLKFNRQCGSTKK | |
| CCSSSSSHHHHHHHHHCHHHHHCCCHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSSSHHHHCCCCHHHHHHHHHHCSSSCCCCCHHHHHHHHCCCSSHHHHHHHHHHHHHHHHHCSSSSSSCCCCCCCSSSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHCHHHCHHHSSCCCCCHHHHHHHHHCCCCCCCHHHHHHHCCCCCCCC | |
| 914874111367861068998428136666523433345789999999999999999899999999999998387999599999999999999999976999999973430589886159998726645310657888886216881898217778772677347699999999998751305899935654440337998851288999998899988999999999999975613154321110000024467889999878989879898999999997529996899985479943201999995013252733332079762189999828778880786876301577789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MLKLVIIENMAEIMLFSLDLLLFSTDILCFNFPSKMIKLPGFITIQIFFYPQASFGISANTILLLFHIFTFVFSHRSKSIDMIISHLSLIHILLLFTQAILVSLDFFGSQNTQDDLRYKVIVFLNKVMRGLSICTPCLLSVLQAIISPSIFSLAKLKHPSASHILGFFLFSWVLNMFIGVIFCCTLRLPPVKRGQSSVCHTALFLFAHELHPQETVFHTNDFEGCHLYRVHGPLKRLHGDYFIQTIRGYLSAFTQPACPRVSPVKRASQAILLLVSFVFTYWVDFTFSFSGGVTWINDSLLVWLQVIVANSYAAISPLMLIYADNQIFKTLQMLWFKYLSPPKLMLKFNRQCGSTKK | |
| 602000033023000000101010131010213342321122000100022113333333320132100000243331110000100131011102011211000000221013331000000000001020000000000100000032120340122002000000010021002010300010203323323210010000000100010111110100000002101200000100211233333343344632333311310000000000000000210000000100334210000000030131010000000001320020020003253421300230354244468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCSSSSSHHHHHHHHHCHHHHHCCCHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSSSHHHHCCCCHHHHHHHHHHCSSSCCCCCHHHHHHHHCCCSSHHHHHHHHHHHHHHHHHCSSSSSSCCCCCCCSSSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHCHHHCHHHSSCCCCCHHHHHHHHHCCCCCCCHHHHHHHCCCCCCCC MLKLVIIENMAEIMLFSLDLLLFSTDILCFNFPSKMIKLPGFITIQIFFYPQASFGISANTILLLFHIFTFVFSHRSKSIDMIISHLSLIHILLLFTQAILVSLDFFGSQNTQDDLRYKVIVFLNKVMRGLSICTPCLLSVLQAIISPSIFSLAKLKHPSASHILGFFLFSWVLNMFIGVIFCCTLRLPPVKRGQSSVCHTALFLFAHELHPQETVFHTNDFEGCHLYRVHGPLKRLHGDYFIQTIRGYLSAFTQPACPRVSPVKRASQAILLLVSFVFTYWVDFTFSFSGGVTWINDSLLVWLQVIVANSYAAISPLMLIYADNQIFKTLQMLWFKYLSPPKLMLKFNRQCGSTKK | |||||||||||||||||||||||||
| 1 | 4grvA | 0.11 | 0.14 | 0.78 | 0.99 | Download | -----------------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR-----KSLQTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAHP----FKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTH-PGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMSGSVQ-----------ALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQWFYHYFYMLTNALAYASPILYNLVSANFRQV--------------------------- | |||||||||||||||||||
| 2 | 4grvA | 0.14 | 0.14 | 0.76 | 1.75 | Download | --------------------------------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLF----TLARKSLQSTVHYHLGSLALSDLLILLL-AMPVELYNFIWVHHFGDAGCRGYYFLRDACTYATALNVASLSVYLAICHP----FKAKTLMSRSRTKKFISAIWLASALLAIPFTMGLQNRSADGTHPGGLVCTPIVTVKVVIQVNTFMSFLFPMLVISILNTVIANKLT-----------------VMSGSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQYFYMLTNALAYASSAINPILYNLVSANFRQ---------------------------- | |||||||||||||||||||
| 3 | 2z73A | 0.10 | 0.17 | 0.91 | 1.23 | Download | ------------ETWWYNPSIVVHPHWREFDQVPD----AVYYSLGIFIGICGIIGCGGNGIVIYLFTKT---KSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMA--ASKKMSHRRAFIMIIFVWLWSVLWAIG--------PIFGWGAYTLEGVLCNCSFDYINILCMFILGFGPILIIFFCYFNIVM-SVSNHEKEMAAMAKRLNAKELAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTWVLTCCQFEDDKDAETEIPAGE- | |||||||||||||||||||
| 4 | 4ib4 | 0.13 | 0.16 | 0.82 | 1.56 | Download | -----------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAV---SLEKKLQYATNYFLMSLAVADLLVGLFV-MPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIKKPIQAN----QYNSRATAFIKITVVWLISIGIAIPVPIKGIETN---PNNITCVLTKFGLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNADAQKGKKNAYIQKYLQTISNEQRASKVLGIVFFFLLMWCPFFITNITLVLCDSTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----------------- | |||||||||||||||||||
| 5 | 3odu | 0.09 | 0.17 | 0.84 | 1.24 | Download | ------------------------------PCFREENANFNKIFLPTIYSIIFLTGIVGNGLVILVM----GYQKLRSMTDKYRLHLSVADLLFVIT--LPFWAVDAVANWYFGNFLCKAVHVIYTVNLYSSVWILAFISLDRYIVHATNSQRP-R---KLLAEKVVYVGVWIPALLLTIPDFIFANVSEAD-------DRYICDRFYDVLPGIVILSCYCIIISKLSHSGSNIFEMLRIDEGKSEKTFRTGTWDAYGSKGHQKRKALKTTVILILAFFACWLPYIGISIDSFILLEVHKWISITEALAFFHCCLNPILYAFLGAKFKTSAQHALTSGRPLEVLFQ----------- | |||||||||||||||||||
| 6 | 4grvA | 0.10 | 0.14 | 0.78 | 1.19 | Download | --------------------------------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR----KSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARYLCHPFKAKT----LMSRSRTKKFISAIWLASALLAIPMLTMGLQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMS-------------GSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEHYFYMLTNALAYASSAINPILYNLVSANFRQV--------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.10 | 0.15 | 0.80 | 1.73 | Download | ----------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFV---IIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDF--RTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECS---LQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNITWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF--------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.14 | 0.17 | 0.82 | 1.72 | Download | ------------------------------GSPRSASSLALAIAITALYSAVCAVGLLGNVLV----MFGIVRYTKMKTTNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFR--TPAKAKLINICIWVLASGVGVPMAVTRPRDG-------AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL-----SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------- | |||||||||||||||||||
| 9 | 4grvA | 0.12 | 0.14 | 0.77 | 1.17 | Download | -----------------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA-----RKSLQTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARAICHPFKAK-----LMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMSGSVQ------------ALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQYFYMLTNALAYASSAINPIL-VSANRQV------------------------------ | |||||||||||||||||||
| 10 | 4ww3A | 0.11 | 0.17 | 0.91 | 1.09 | Download | -----------ETWWYNPSIVVH---PHWREF--DQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTK---TKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWFGFAACKVYGFIGGIFGFMSIMTMAMISIDRVIGRPMAASKKMSHRRA----FIMIIFVWLWSVLWAIGPIFYTCSFDYISRDSTTRSNILCMFILGFFGILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGAN--------AEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVPYAAQLPVMFAKASAIPMIYSVSHPKFREAISQTWVLTCCQFDDKETEDDKDAETEI | |||||||||||||||||||
| ||||||||||||||||||||||||||
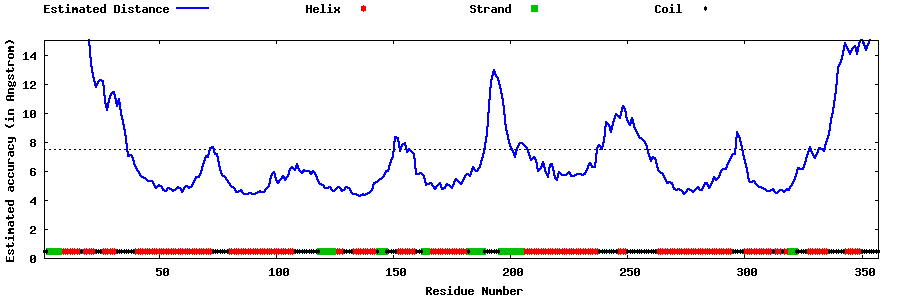
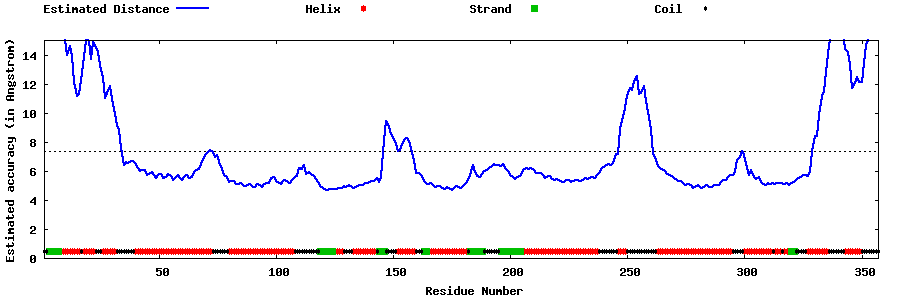
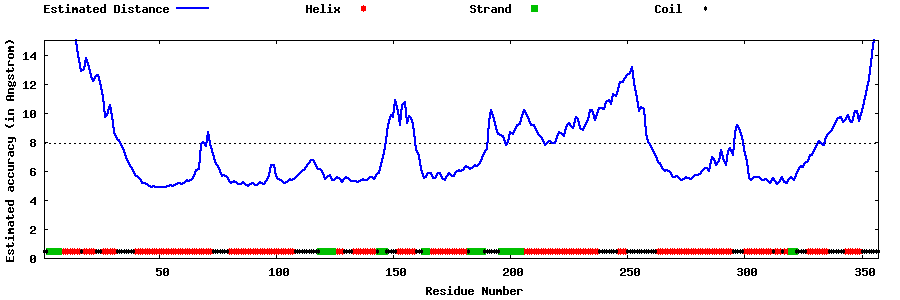
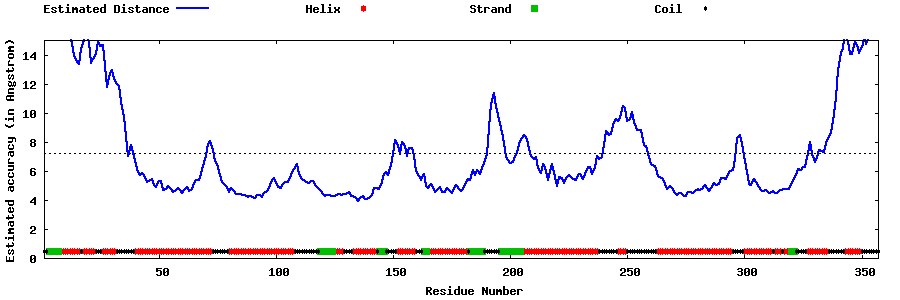
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||