

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MATELDKIFLILAIAEFIISMLGNVFIGLVNCSEGIKNQKVFSADFILTCLAISTIGQLLVILFDSFLVGLASHLYTTYRLGKTVIMLWHMTNHLTTWLATCLSIFYFFKIAHFPHSLFLWLRWRMNGMIVMLLILSLFLLIFDSLVLEIFIDISLNIIDKSNLTLYLDESKTLYDKLSILKTLLSLTSFIPFSLFLTSLLFLFLSLVRHTRNLKLSSLGSRDSSTEAHRRAMKMVMSFLFLFIVHFFSLQVANGIFFMLWNNKYIKFVMLALNAFPSCHSFILILGNSKLRQTAVRLLWHLRNYTKTPNALPL | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHSSCSSSSSSCCCSSSSSSSSSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCSSSCCCCCCC | |
| 98689999999999999999999899999999999808989868899999999999999999985754774253113578999999999998299999999999998567607997899998987438098999999999999999998230116520147512368887576454069999999999999999999999999999999999973768999999808999999999999999999999999999998705760999999999999688877988438769999999999828286418987899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MATELDKIFLILAIAEFIISMLGNVFIGLVNCSEGIKNQKVFSADFILTCLAISTIGQLLVILFDSFLVGLASHLYTTYRLGKTVIMLWHMTNHLTTWLATCLSIFYFFKIAHFPHSLFLWLRWRMNGMIVMLLILSLFLLIFDSLVLEIFIDISLNIIDKSNLTLYLDESKTLYDKLSILKTLLSLTSFIPFSLFLTSLLFLFLSLVRHTRNLKLSSLGSRDSSTEAHRRAMKMVMSFLFLFIVHFFSLQVANGIFFMLWNNKYIKFVMLALNAFPSCHSFILILGNSKLRQTAVRLLWHLRNYTKTPNALPL | |
| 74332332113113312331331123002100110045340310110000002110000001232100000003113332000001010112013103000300100000002231100000122033200110131223223111111031313243334322223133333333210011123333333133323323210010022023202433544411314002200100120221133213112311011233432211210123101000010204314143013001200210202245663247 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHSSCSSSSSSCCCSSSSSSSSSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCSSSCCCCCCC MATELDKIFLILAIAEFIISMLGNVFIGLVNCSEGIKNQKVFSADFILTCLAISTIGQLLVILFDSFLVGLASHLYTTYRLGKTVIMLWHMTNHLTTWLATCLSIFYFFKIAHFPHSLFLWLRWRMNGMIVMLLILSLFLLIFDSLVLEIFIDISLNIIDKSNLTLYLDESKTLYDKLSILKTLLSLTSFIPFSLFLTSLLFLFLSLVRHTRNLKLSSLGSRDSSTEAHRRAMKMVMSFLFLFIVHFFSLQVANGIFFMLWNNKYIKFVMLALNAFPSCHSFILILGNSKLRQTAVRLLWHLRNYTKTPNALPL | |||||||||||||||||||||||||
| 1 | 4djhA | 0.12 | 0.18 | 0.89 | 1.36 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVL-GGTKVREDVDVIECSLQFDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL------------DRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 2 | 5tjvA | 0.09 | 0.18 | 0.88 | 2.61 | Download | LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS--RSLRCRPSYHFIGSLAVADLLGSVIFVYSFID-FHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPK----AVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDI---------FPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.12 | 0.20 | 0.90 | 2.35 | Download | SSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMK-TATNIYIFNLALADALATST--LPFQSAKYLMETWPFELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPAKAKLINICIWVLASGVGVPIMVMA-----VTRPRDGAVVCMLQPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV-----RLLSGSK-EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------- | |||||||||||||||||||
| 4 | 3uon | 0.11 | 0.22 | 0.89 | 1.59 | Download | --TFEVVFIVLVAGSLSLVTIIGNILVMVSIKV---NRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPL-GPVVCLWLALDYVVSNASVMNLLIISFDRYFCVT---KPLTYP-VKRTTKMAGMMIAAAWVLFILWAPAILFWQFIVVDGECYIQFF-----SNAAVTF-----GTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMGGAADAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------- | |||||||||||||||||||
| 5 | 5glh | 0.13 | 0.23 | 0.88 | 1.20 | Download | IKETFKYINTVVSCLVFVLGIIGNSTLLYIIYK--------NGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASW--------------WTAVEIVLIWVVSVVLAVPEAIGFDIITMDY-KGSYLRICLLHPVQKAFMQFYWWLFSFYFCLPLAITAFFYTLMTCEMLRKNEGLRLTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKTLYNQLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL--------------- | |||||||||||||||||||
| 6 | 4djhA | 0.11 | 0.18 | 0.89 | 1.52 | Download | --PAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY---TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKAL----DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 7 | 3uon | 0.09 | 0.22 | 0.90 | 1.72 | Download | -----VVFIVLVAGSLSLVTIIGNILVMVSIKV---NRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRT--VEDGECYIQ----FFS--NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMTAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP-CIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------- | |||||||||||||||||||
| 8 | 4buoA | 0.12 | 0.19 | 0.89 | 2.59 | Download | TDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR----KKSLSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICH-------PFKAKTRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHGLVCTPIV----DTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQP------GRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL--------------- | |||||||||||||||||||
| 9 | 4djhA | 0.11 | 0.18 | 0.89 | 1.53 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKA-LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL------------DRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAASSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 10 | 1l9hA | 0.10 | 0.22 | 0.94 | 1.64 | Download | AEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQ----HKKLTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCPMSNFRFG------ENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTV----------KEAAAATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDSTTVSK | |||||||||||||||||||
| ||||||||||||||||||||||||||
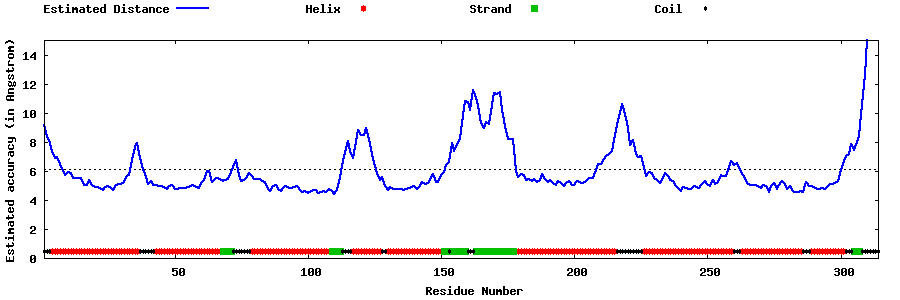
| Generated 3D models | Estimated local accuracy of models | ||
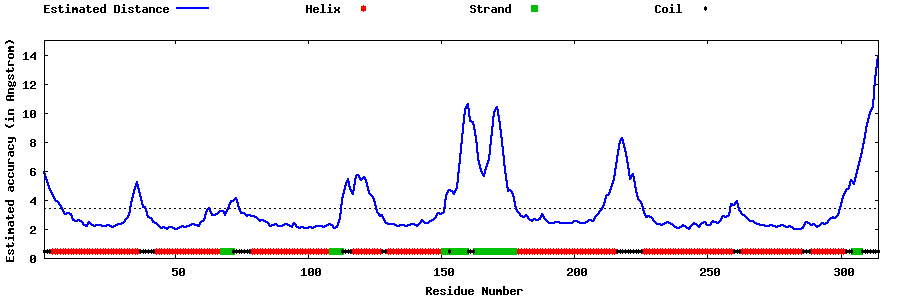 | |||
|
|
|||
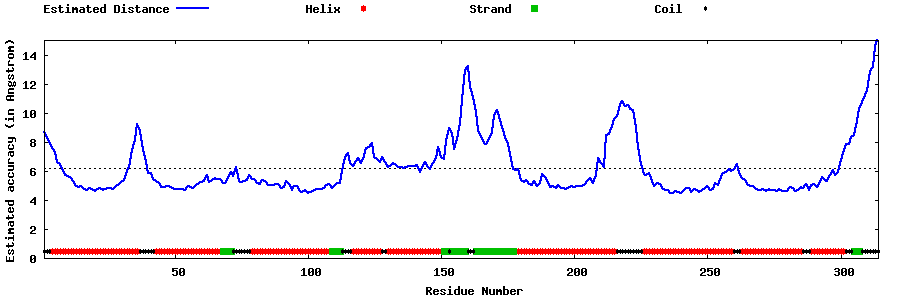 | |||
|
| |||
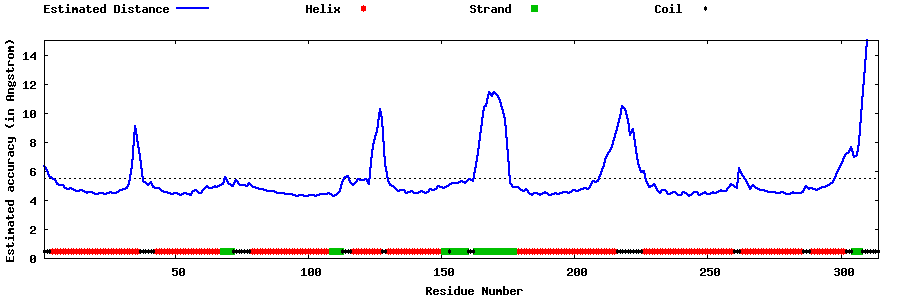 | |||
|
| |||
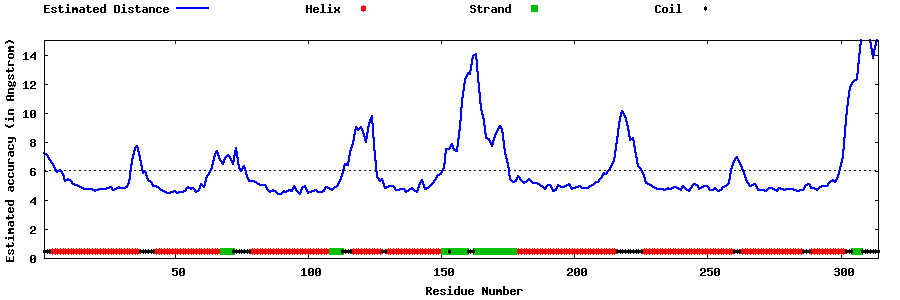 | |||
|
| |||
 | |||
|
| |||