

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MPANFTEGSFDSSGTGQTLDSSPVACTETVTFTEVVEGKEWGSFYYSFKTEQLITLWVLFVFTIVGNSVVLFSTWRRKKKSRMTFFVTQLAITDSFTGLVNILTDINWRFTGDFTAPDLVCRVVRYLQVVLLYASTYVLVSLSIDRYHAIVYPMKFLQGEKQARVLIVIAWSLSFLFSIPTLIIFGKRTLSNGEVQCWALWPDDSYWTPYMTIVAFLVYFIPLTIISIMYGIVIRTIWIKSKTYETVISNCSDGKLCSSYNRGLISKAKIKAIKYSIIIILAFICCWSPYFLFDILDNFNLLPDTQERFYASVIIQNLPALNSAINPLIYCVFSSSISFPCREQRSQDSRMTFRERTERHEMQILSKPEFI | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCSSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 98998999888888888888898788887777766666555553257999999999999999999999999999747888703999999999999999998799999999488363245885299999999999999999997011412135687411388999998999999999999999987759936990799861897538999999999999999999999999999999976258654321222323222310142788988899999999999999998999999999997355464167999999999999999899999998199999999998466567888888877778757887789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MPANFTEGSFDSSGTGQTLDSSPVACTETVTFTEVVEGKEWGSFYYSFKTEQLITLWVLFVFTIVGNSVVLFSTWRRKKKSRMTFFVTQLAITDSFTGLVNILTDINWRFTGDFTAPDLVCRVVRYLQVVLLYASTYVLVSLSIDRYHAIVYPMKFLQGEKQARVLIVIAWSLSFLFSIPTLIIFGKRTLSNGEVQCWALWPDDSYWTPYMTIVAFLVYFIPLTIISIMYGIVIRTIWIKSKTYETVISNCSDGKLCSSYNRGLISKAKIKAIKYSIIIILAFICCWSPYFLFDILDNFNLLPDTQERFYASVIIQNLPALNSAINPLIYCVFSSSISFPCREQRSQDSRMTFRERTERHEMQILSKPEFI | |
| 74443343313343333313122232233233343343343332222020000211110021013000000000013323010000010003001010000010100000143220120001003001000000000000000130020001002312133100000000010000000000000002426643212113012630210000000121133102101300020001013334335444544454443444444323301100000000000000000221000000100131243412100100001000200021000000004300410240032344343465344443533345624 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCSSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCC MPANFTEGSFDSSGTGQTLDSSPVACTETVTFTEVVEGKEWGSFYYSFKTEQLITLWVLFVFTIVGNSVVLFSTWRRKKKSRMTFFVTQLAITDSFTGLVNILTDINWRFTGDFTAPDLVCRVVRYLQVVLLYASTYVLVSLSIDRYHAIVYPMKFLQGEKQARVLIVIAWSLSFLFSIPTLIIFGKRTLSNGEVQCWALWPDDSYWTPYMTIVAFLVYFIPLTIISIMYGIVIRTIWIKSKTYETVISNCSDGKLCSSYNRGLISKAKIKAIKYSIIIILAFICCWSPYFLFDILDNFNLLPDTQERFYASVIIQNLPALNSAINPLIYCVFSSSISFPCREQRSQDSRMTFRERTERHEMQILSKPEFI | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.18 | 0.20 | 0.80 | 2.75 | Download | ------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.20 | 0.23 | 0.85 | 3.88 | Download | ---------------------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKMRNGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDYKLRICLLHPVQKFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNIFEMLRIDEGGGSGGDEAEKLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC------------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.18 | 0.20 | 0.80 | 3.65 | Download | ------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSK----------------EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 4 | 4djh | 0.21 | 0.23 | 0.80 | 1.54 | Download | --------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYWLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFDEGLNGKRNQTPAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA--A---LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------ | |||||||||||||||||||
| 5 | 4rnb | 0.21 | 0.22 | 0.81 | 1.16 | Download | --------------------------------------------PKEYEWVLIAGYIIVFVVALIGNVLVCVAVWKNHMRTVTNYFIVNLSLADVLVTITCLPATLVVDITETWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPS----TAKRARNSIVIIWIVSCIIMIPQAIVMECSTVFTLFTVCDERWGGEIYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSFWNKKRQSRSDRAMSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMHDRETVYAWFTFSHWLVYANSAANPIIYNFLSGKFREEFKAAFSC------------------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.18 | 0.20 | 0.79 | 3.01 | Download | ---------------------------------------ARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 7 | 3uon | 0.21 | 0.23 | 0.79 | 1.72 | Download | ------------------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVVEDGECYIQFFS---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDERNGVRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCI---PNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.20 | 0.19 | 0.74 | 4.06 | Download | --------------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPT-----SSKAQAVNVAIWALASVVGVPVAIMGS-AQVEDEEIECLVETPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS----------------REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR----------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.18 | 0.20 | 0.80 | 2.82 | Download | ------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.19 | 0.19 | 0.78 | 3.36 | Download | ---------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR-QGSIDCTLTFSHWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS----------------KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
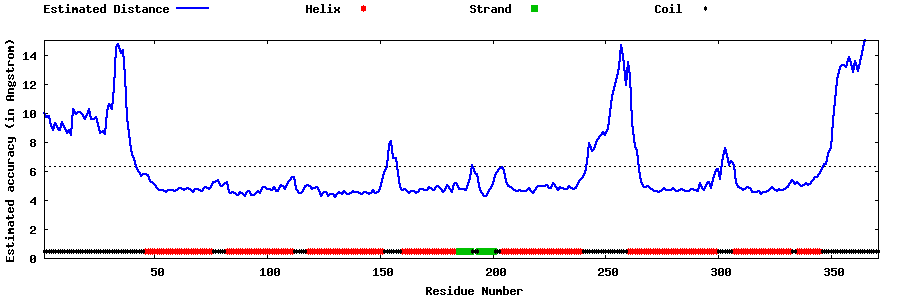
| Generated 3D models | Estimated local accuracy of models | ||
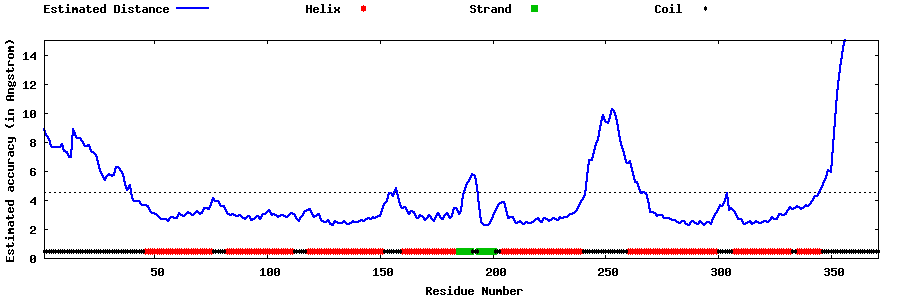 | |||
|
|
|||
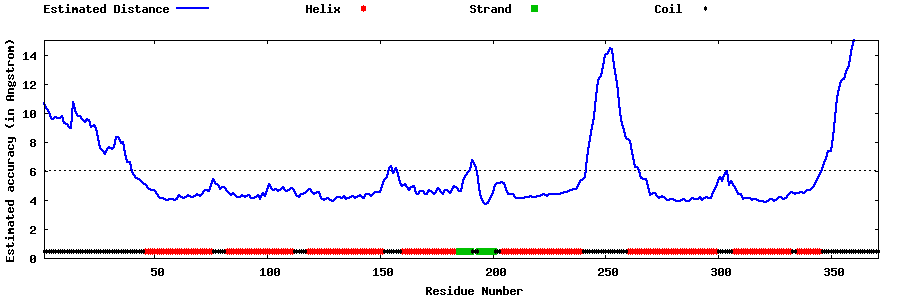 | |||
|
| |||
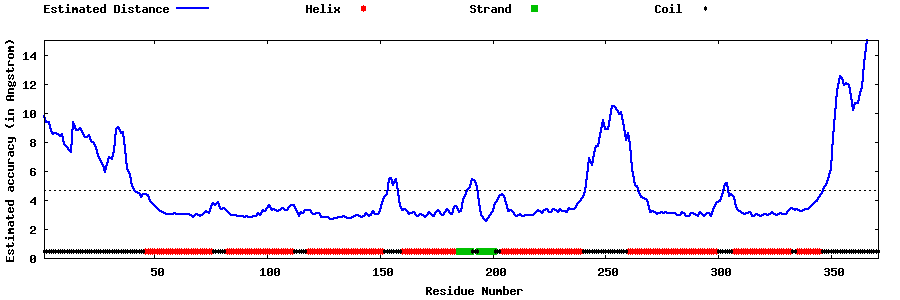 | |||
|
| |||
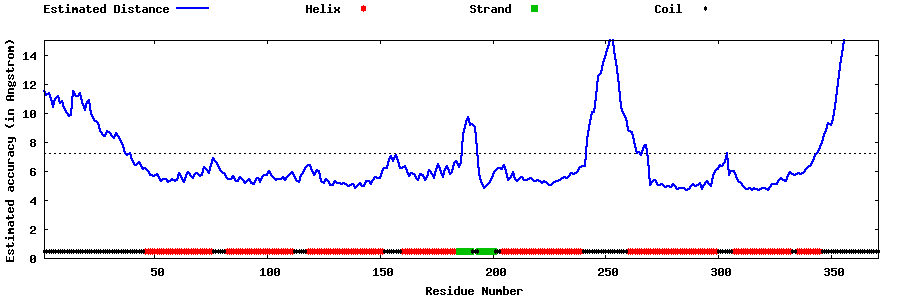 | |||
|
| |||
 | |||
|
| |||