

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MAETLQLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIYLLALLGNGALPAVVWIDSTLHQPMFLLLAILAATDLGLATSIAPGLLAVLWLGPRSVPYAVCLVQMFFVHALTAMESGVLLAMACDRAAAIGRPLHYPVLVTKACVGYAALALALKAVAIVVPFPLLVAKFEHFQAKTIGHTYCAHMAVVELVVGNTQATNLYGLALSLAISGMDILGITGSYGLIAHAVLQLPTREAHAKAFGTCSSHICVILAFYIPGLFSYLTHRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGARTKQIRDRLLETFTFRKSPL | |
| CCCCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHHCCCCC | |
| 96402688999857411477899987889999999999999999999998887747887543289999999999999999539999999956998678888999999999999999999999981327862666556501788899999999999999999889999952899998967875655287788836794378899999999999998999999999999998179898898998650699999999999999998011024799996089999999996278664680211657999999999974356899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MAETLQLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIYLLALLGNGALPAVVWIDSTLHQPMFLLLAILAATDLGLATSIAPGLLAVLWLGPRSVPYAVCLVQMFFVHALTAMESGVLLAMACDRAAAIGRPLHYPVLVTKACVGYAALALALKAVAIVVPFPLLVAKFEHFQAKTIGHTYCAHMAVVELVVGNTQATNLYGLALSLAISGMDILGITGSYGLIAHAVLQLPTREAHAKAFGTCSSHICVILAFYIPGLFSYLTHRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGARTKQIRDRLLETFTFRKSPL | |
| 64403331313230310200000121320200011022012203321210000021141001000200010010000002000020000000203403040000001200310230110011003000000021010000002310010000002212011202102021022044200000001021001000230300111001003313332231032011200200010025601320120010010001003221331010000043300210000002310030033003000020340041003002445668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHHCCCCC MAETLQLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIYLLALLGNGALPAVVWIDSTLHQPMFLLLAILAATDLGLATSIAPGLLAVLWLGPRSVPYAVCLVQMFFVHALTAMESGVLLAMACDRAAAIGRPLHYPVLVTKACVGYAALALALKAVAIVVPFPLLVAKFEHFQAKTIGHTYCAHMAVVELVVGNTQATNLYGLALSLAISGMDILGITGSYGLIAHAVLQLPTREAHAKAFGTCSSHICVILAFYIPGLFSYLTHRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGARTKQIRDRLLETFTFRKSPL | |||||||||||||||||||||||||
| 1 | 3emlA | 0.21 | 0.24 | 0.86 | 2.36 | Download | ------------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHRQ-- | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.26 | 0.84 | 2.10 | Download | --------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 3 | 5tgzA | 0.20 | 0.26 | 0.88 | 1.72 | Download | --------GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 4 | 4ib4 | 0.18 | 0.20 | 0.88 | 1.51 | Download | ----------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIE-T-----NPNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAAISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR-- | |||||||||||||||||||
| 5 | 3uonA | 0.16 | 0.19 | 0.85 | 1.13 | Download | -------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFI--VGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNT-VWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------ | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.24 | 0.87 | 2.57 | Download | --------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVAC-----------LFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.23 | 0.82 | 1.69 | Download | --------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQA----------SECVV----------NTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH----L-AIFDF-FTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK---- | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.20 | 0.89 | 2.81 | Download | -------NSDL---------DVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGI-----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL------- | |||||||||||||||||||
| 9 | 5tgzA | 0.19 | 0.26 | 0.89 | 3.08 | Download | --GGRGENFMDIECFMVL-------PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 10 | 4gpoA | 0.17 | 0.20 | 0.87 | 4.54 | Download | ------------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP-DWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL-------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
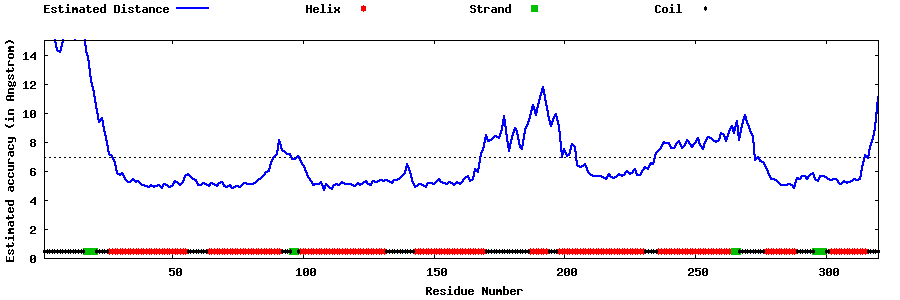
| Generated 3D models | Estimated local accuracy of models | ||
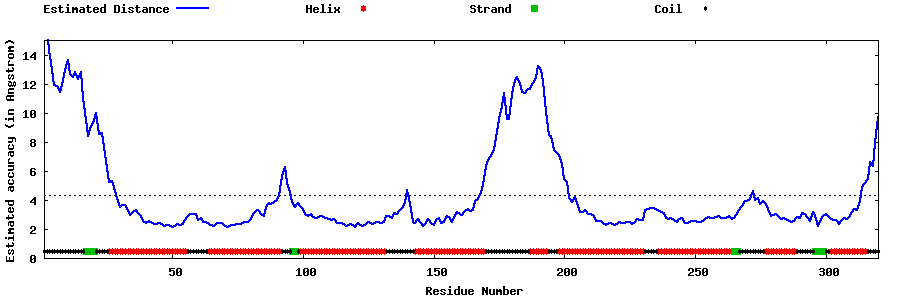 | |||
|
|
|||
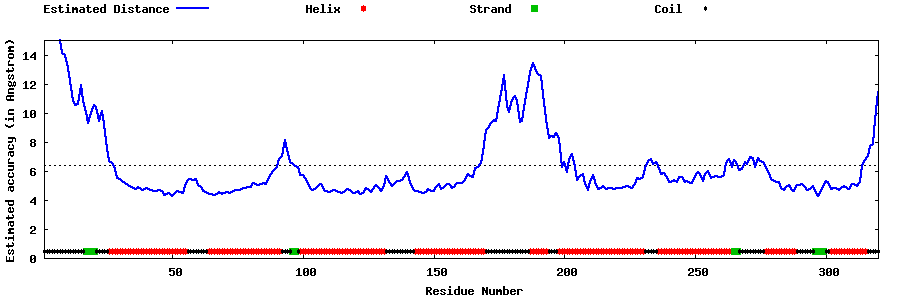 | |||
|
| |||
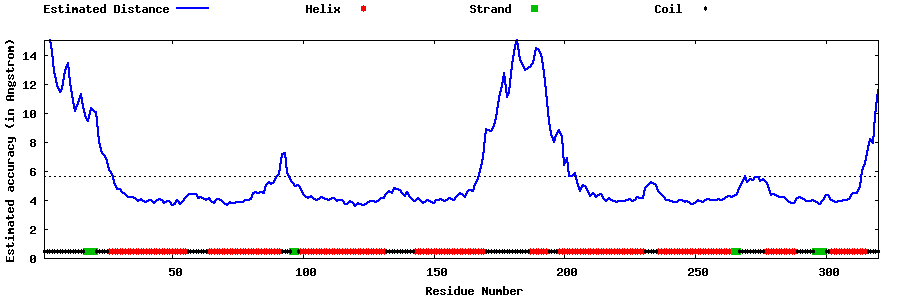 | |||
|
| |||
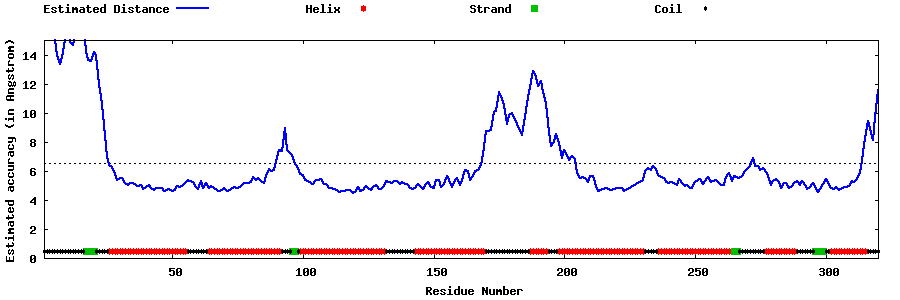 | |||
|
| |||
 | |||
|
| |||