

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MEHTHAHLAANSSLSWWSPGSACGLGFVPVVYYSLLLCLGLPANILTVIILSQLVARRQKSSYNYLLALAAADILVLFFIVFVDFLLEDFILNMQMPQVPDKIIEVLEFSSIHTSIWITVPLTIDRYIAVCHPLKYHTVSYPARTRKVIVSVYITCFLTSIPYYWWPNIWTEDYISTSVHHVLIWIHCFTVYLVPCSIFFILNSIIVYKLRRKSNFRLRGYSTGKTTAILFTITSIFATLWAPRIIMILYHLYGAPIQNRWLVHIMSDIANMLALLNTAINFFLYCFISKRFRTMAAATLKAFFKCQKQPVQFYTNHNFSITSSPWISPANSHCIKMLVYQYDKNGKPIKVSP | |
| CCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCCCCCCCCCCCCC | |
| 99988888899887888888760175599999999989988768854014344698876750999999999999999997559999999666972733389999999999999999999999998667643007041114486789878899999999984998771589667888406569999999999999999999999999999843567430012026787888999999999989999999999972354623699999999999999999899999999299999999999988526567887667777877555788777998843201356888998466898 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MEHTHAHLAANSSLSWWSPGSACGLGFVPVVYYSLLLCLGLPANILTVIILSQLVARRQKSSYNYLLALAAADILVLFFIVFVDFLLEDFILNMQMPQVPDKIIEVLEFSSIHTSIWITVPLTIDRYIAVCHPLKYHTVSYPARTRKVIVSVYITCFLTSIPYYWWPNIWTEDYISTSVHHVLIWIHCFTVYLVPCSIFFILNSIIVYKLRRKSNFRLRGYSTGKTTAILFTITSIFATLWAPRIIMILYHLYGAPIQNRWLVHIMSDIANMLALLNTAINFFLYCFISKRFRTMAAATLKAFFKCQKQPVQFYTNHNFSITSSPWISPANSHCIKMLVYQYDKNGKPIKVSP | |
| 65344342331322333433221000000121011002102311200000000034133302110010001001020001021000001113301012002100000110001000000000000001000100202321223200000000000000001110000002336323300210000110211112101000200010001024355464444431100000000000000012220010001001342323200000101010000200023000100004301410240033014144444633444433333344434444433433345454444335347 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCCCCCCCCCCCCC MEHTHAHLAANSSLSWWSPGSACGLGFVPVVYYSLLLCLGLPANILTVIILSQLVARRQKSSYNYLLALAAADILVLFFIVFVDFLLEDFILNMQMPQVPDKIIEVLEFSSIHTSIWITVPLTIDRYIAVCHPLKYHTVSYPARTRKVIVSVYITCFLTSIPYYWWPNIWTEDYISTSVHHVLIWIHCFTVYLVPCSIFFILNSIIVYKLRRKSNFRLRGYSTGKTTAILFTITSIFATLWAPRIIMILYHLYGAPIQNRWLVHIMSDIANMLALLNTAINFFLYCFISKRFRTMAAATLKAFFKCQKQPVQFYTNHNFSITSSPWISPANSHCIKMLVYQYDKNGKPIKVSP | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.20 | 0.19 | 0.81 | 2.97 | Download | -------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTA-TNIYIFNLALADALATSTLPFQSAKYLM---TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP-------------------------------------------------- | |||||||||||||||||||
| 2 | 4bwbA | 0.23 | 0.22 | 0.80 | 3.67 | Download | --------------SDLDVNTDIYSKVLVTAIYLALFVVGTVGNGVTLFTL--AQSLQSRVD-YYLGSLALSDLLILLFALPVDLYNFIWHHPWAFGDAGCKGYYFLREACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSIVDTATLRVVIQLNTFMSFLFPMLVASILNTVAARRLTVMVEPG-RVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFVYISDEQWTDFYHYFYMLSNALVYVSAAINPILYNLVSANFRQVFLSTLAC--------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.19 | 0.19 | 0.82 | 3.45 | Download | -------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTA-TNIYIFNLALADALATSTLPFQSAKYLME--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 4 | 4ib4 | 0.16 | 0.21 | 0.80 | 1.56 | Download | -------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSL--EKKLQYATNFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLLVLCP-AWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGITNPNNITCVLTKEDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN-QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------------------------------------------ | |||||||||||||||||||
| 5 | 4djh | 0.18 | 0.23 | 0.77 | 1.16 | Download | ---------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATN-IYIFNLALADALVTTT-MPFQST-VYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTCSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---A---LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.20 | 0.19 | 0.81 | 3.20 | Download | ---------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATN-IYIFNLALADALATSTLPFQSAKYLME--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 7 | 4ib4 | 0.17 | 0.21 | 0.78 | 1.72 | Download | -------------------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQ-YATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC-NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCN-------------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.20 | 0.19 | 0.82 | 3.85 | Download | ------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKT-ATNIYIFNLALADALATSTLPFQSAKYLME--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK--PCG---------------------------------------------- | |||||||||||||||||||
| 9 | 5o9hA | 0.15 | 0.15 | 0.80 | 3.11 | Download | -------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTA-FEAKRTIN-AIWFLNLAVADFLACLALPALFTSIVQ--HHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETR-----STKTLKVVVAVVASFFIFWLPYQVTGIMMSFLESSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------------------------ | |||||||||||||||||||
| 10 | 4n6hA | 0.19 | 0.19 | 0.86 | 3.62 | Download | LKTTNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTA-TNIYIFNLALADALATSTLPFQSAKYLM--ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
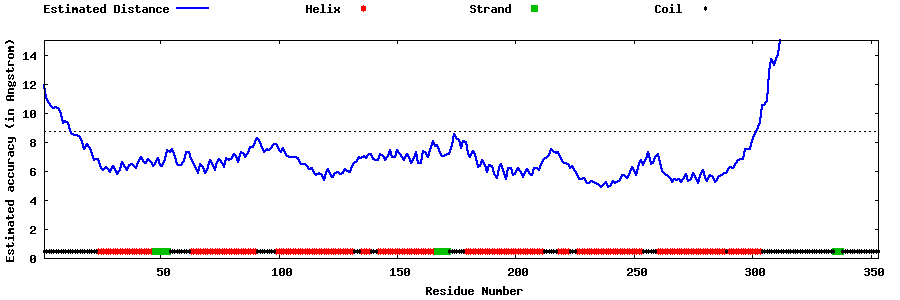
| Generated 3D models | Estimated local accuracy of models | ||
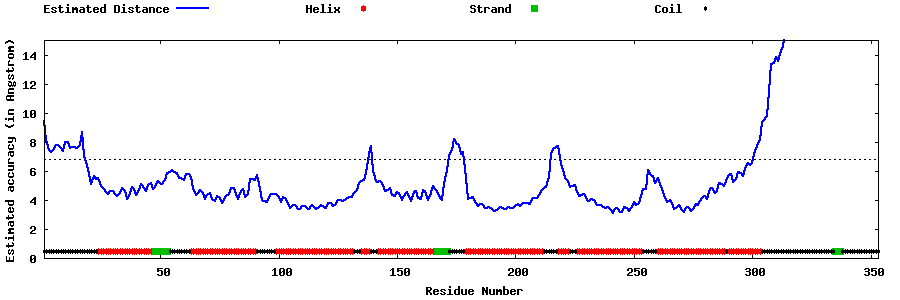 | |||
|
|
|||
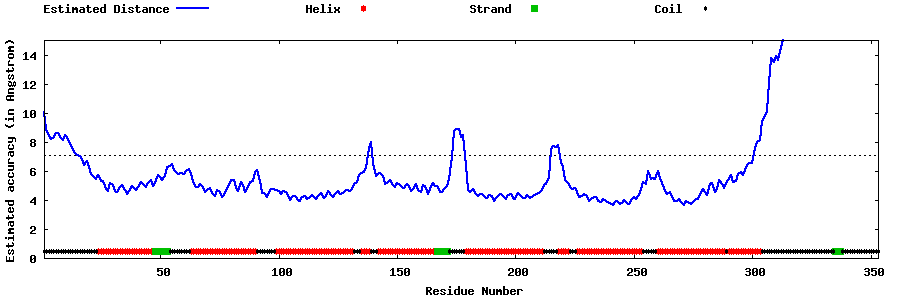 | |||
|
| |||
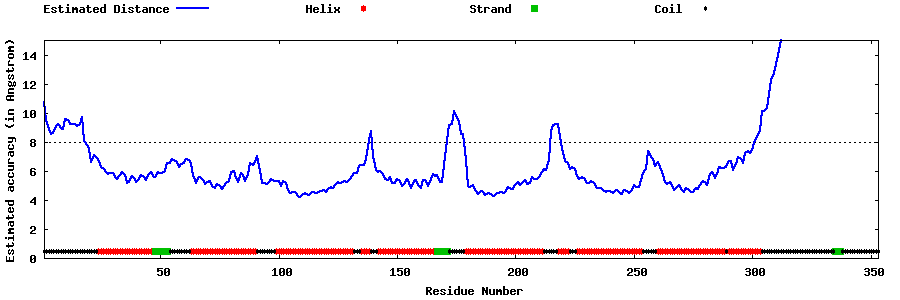 | |||
|
| |||
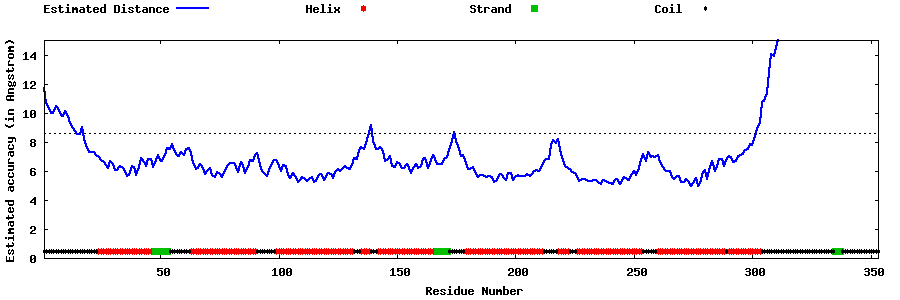 | |||
|
| |||
 | |||
|
| |||