

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSPECARAAGDAPLRSLEQANRTRFPFFSDVKGDHRLVLAAVETTVLVLIFAVSLLGNVCALVLVARRRRRGATACLVLNLFCADLLFISAIPLVLAVRWTEAWLLGPVACHLLFYVMTLSGSVTILTLAAVSLERMVCIVHLQRGVRGPGRRARAVLLALIWGYSAVAALPLCVFFRVVPQRLPGADQEISICTLIWPTIPGEISWDVSFVTLNFLVPGLVIVISYSKILQTSEHLLDARAVVTHSEITKASRKRLTVSLAYSESHQIRVSQQDFRLFRTLFLLMVSFFIMWSPIIITILLILIQNFKQDLVIWPSLFFWVVAFTFANSALNPILYNMTLCRNEWKKIFCCFWFPEKGAILTDTSVKRNDLSIISG | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCCCSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 98998889999998877778789898765557885439999999999999999999998742003689998587899999999999999988999999991864280688858999999999999999999999707661035575545468989997799999999999999998367650147889987266321898038899999999999999999999999999999987521222211124556654024556544320367888888899999999999999999999999999997275443118999999999999999999799974188889999985766778888877777777898784889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSPECARAAGDAPLRSLEQANRTRFPFFSDVKGDHRLVLAAVETTVLVLIFAVSLLGNVCALVLVARRRRRGATACLVLNLFCADLLFISAIPLVLAVRWTEAWLLGPVACHLLFYVMTLSGSVTILTLAAVSLERMVCIVHLQRGVRGPGRRARAVLLALIWGYSAVAALPLCVFFRVVPQRLPGADQEISICTLIWPTIPGEISWDVSFVTLNFLVPGLVIVISYSKILQTSEHLLDARAVVTHSEITKASRKRLTVSLAYSESHQIRVSQQDFRLFRTLFLLMVSFFIMWSPIIITILLILIQNFKQDLVIWPSLFFWVVAFTFANSALNPILYNMTLCRNEWKKIFCCFWFPEKGAILTDTSVKRNDLSIISG | |
| 85162453333231332433131323213324343200010010211020023133321000000013301100100020111000000101101000002343001200010100001000000000000001320000010020333233210000000001200100111000010233414444431110103012430320000010332133111000200010011023324444344444344444443434444344444443434100000000000000000222100000100132234320110010100100020000000000012014101200201122553534443434334324248 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCCCSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCC MSPECARAAGDAPLRSLEQANRTRFPFFSDVKGDHRLVLAAVETTVLVLIFAVSLLGNVCALVLVARRRRRGATACLVLNLFCADLLFISAIPLVLAVRWTEAWLLGPVACHLLFYVMTLSGSVTILTLAAVSLERMVCIVHLQRGVRGPGRRARAVLLALIWGYSAVAALPLCVFFRVVPQRLPGADQEISICTLIWPTIPGEISWDVSFVTLNFLVPGLVIVISYSKILQTSEHLLDARAVVTHSEITKASRKRLTVSLAYSESHQIRVSQQDFRLFRTLFLLMVSFFIMWSPIIITILLILIQNFKQDLVIWPSLFFWVVAFTFANSALNPILYNMTLCRNEWKKIFCCFWFPEKGAILTDTSVKRNDLSIISG | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.22 | 0.25 | 0.86 | 2.67 | Download | LANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------GAVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV-------------------------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLD--ENFKRCFRQLCRKPCG------------------ | |||||||||||||||||||
| 2 | 5gliA | 0.21 | 0.24 | 0.86 | 3.85 | Download | ------------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKMRNGPNILIASLALGDLLHVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDY--KGSYLRICLLHPVQKFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRIDEGGGSGGDEAEKLFNQDVDAARTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDLSFLLVLDYIGINMASLNSCANPIALYLVSFKNAFKSALCC------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.23 | 0.79 | 3.41 | Download | ---------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------GAVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL-------------------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLNFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 4 | 4djh | 0.20 | 0.23 | 0.81 | 1.54 | Download | -----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE----DVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEGLRLKTTNGVIQQKRNQTWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLNFKRCFRDFCFP------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.19 | 0.23 | 0.81 | 1.18 | Download | -----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRED----VDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEMLRIDEGLRNQKRNQTGTWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDFKRCFRDFCFP------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.22 | 0.23 | 0.78 | 3.14 | Download | -----------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL-------------------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLNFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 7 | 4djh | 0.20 | 0.23 | 0.81 | 1.74 | Download | -------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE----DVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEMEGLRNTNGVITLQQKQTPDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---A---LSSYYFCIALGYTNSSLNPILYAFLNFKRCFRDFCF-------------------------- | |||||||||||||||||||
| 8 | 4djhA | 0.20 | 0.23 | 0.80 | 4.39 | Download | -----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE----DVDVIECSLQFPDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSQTPNRAKRVITTFRTGTWDAYREKD------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA------LSSYYFCIALGYTNSSLNPILYAFLDFKRCFRDFCF----P--------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.22 | 0.23 | 0.79 | 2.92 | Download | ---------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------GAVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-------------------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLD--ENFKRCF-CRKPCG-------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.20 | 0.19 | 0.77 | 3.50 | Download | ------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQ------GSIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLK-------------------------SVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-TFQTVSWHFCIALGYTNSCLNPVLYAFENFKRCF------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
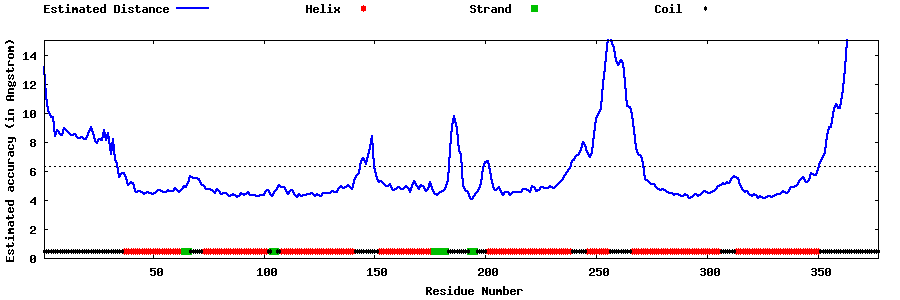
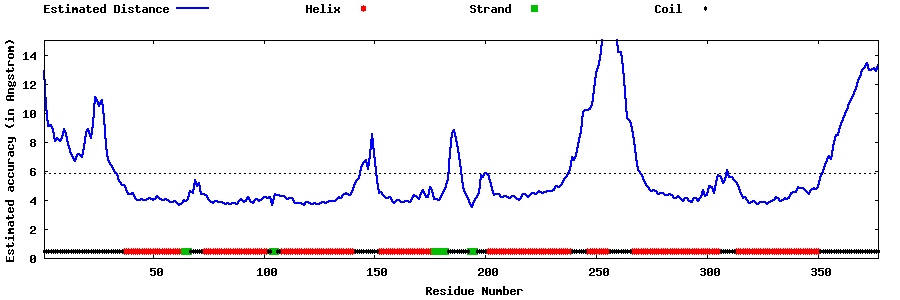
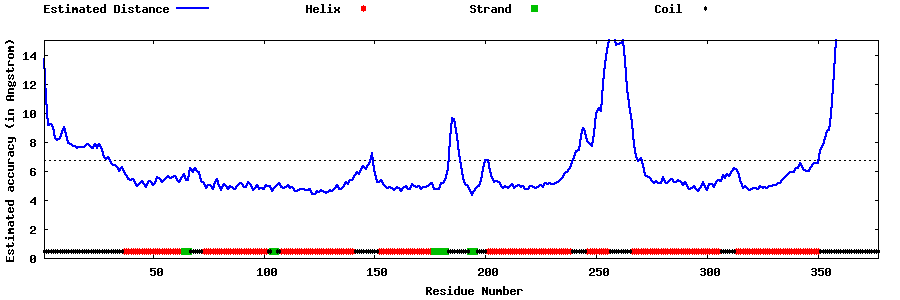
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||