

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDLINSTDYLINASTLVRNSTQFLAPASKMIIALSLYISSIIGTITNGLYLWVLRFKMKQTVNTLLFFHLILSYFISTMILPFMATSQLQDNHWNFGTALCKVFNGTLSLGMFTSVFFLSAIGLDRYLLTLHPVWSQQHRTPRWASSIVLGVWISAAALSIPYLIFRETHHDRKGKVTCQNNYAVSTNWESKEMQASRQWIHVACFISRFLLGFLLPFFIIIFCYERVASKVKERSLFKSSKPFKVMMTAIISFFVCWMPYHIHQGLLLTTNQSLLLELTLILTVLTTSFNTIFSPTLYLFVGENFKKVFKKSILALFESTFSEDSSVERTQT | |
| CCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCSSSSCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCC | |
| 998657878888876667766554369999999999999999999899999988756677789999999999999999999999999977997989628999999999999999999999999999999981255421346378988889999999999989999872066689957882157764354421000367899999999999999999999999999999999974666346758999999999999996899999999997651699999999999999999828489799438889899999989876887788888888999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDLINSTDYLINASTLVRNSTQFLAPASKMIIALSLYISSIIGTITNGLYLWVLRFKMKQTVNTLLFFHLILSYFISTMILPFMATSQLQDNHWNFGTALCKVFNGTLSLGMFTSVFFLSAIGLDRYLLTLHPVWSQQHRTPRWASSIVLGVWISAAALSIPYLIFRETHHDRKGKVTCQNNYAVSTNWESKEMQASRQWIHVACFISRFLLGFLLPFFIIIFCYERVASKVKERSLFKSSKPFKVMMTAIISFFVCWMPYHIHQGLLLTTNQSLLLELTLILTVLTTSFNTIFSPTLYLFVGENFKKVFKKSILALFESTFSEDSSVERTQT | |
| 634403131323443324344422330010000220220023033212200000202332100000000001030100000001000002443210010002200221331111100000000300010000002023223211010000001100000010100002024357232201020234332444334333301200111111110133112101300010011023264464300000000000001103231200000000043331010000000020134013110000000340142014002410431045554465568 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCSSSSCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCC MDLINSTDYLINASTLVRNSTQFLAPASKMIIALSLYISSIIGTITNGLYLWVLRFKMKQTVNTLLFFHLILSYFISTMILPFMATSQLQDNHWNFGTALCKVFNGTLSLGMFTSVFFLSAIGLDRYLLTLHPVWSQQHRTPRWASSIVLGVWISAAALSIPYLIFRETHHDRKGKVTCQNNYAVSTNWESKEMQASRQWIHVACFISRFLLGFLLPFFIIIFCYERVASKVKERSLFKSSKPFKVMMTAIISFFVCWMPYHIHQGLLLTTNQSLLLELTLILTVLTTSFNTIFSPTLYLFVGENFKKVFKKSILALFESTFSEDSSVERTQT | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.28 | 0.26 | 0.87 | 3.25 | Download | ----------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYPPKVLCGVDHD--------------KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVL----TEESVVR---- | |||||||||||||||||||
| 2 | 4xnwA | 0.19 | 0.22 | 0.86 | 4.27 | Download | ----------------SFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTS-----------DEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR--------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.26 | 0.87 | 3.88 | Download | ----------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKKTATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSP-----------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.18 | 0.20 | 0.84 | 1.51 | Download | ----------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVLEKKLQYATNYFLMSLAVADLLVGLFMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGI-ET-NPNNITCVLTKER----------------FGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAASNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR-------------- | |||||||||||||||||||
| 5 | 4djh | 0.26 | 0.24 | 0.83 | 1.17 | Download | ------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDD----------YSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA---LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.27 | 0.26 | 0.86 | 3.42 | Download | -----------------------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYPPKVLCGVDHD--------------KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR-------- | |||||||||||||||||||
| 7 | 4djh | 0.26 | 0.24 | 0.83 | 1.69 | Download | -------------------------PAIPVIITAVYSVVFVVGLVGNSLVMFVIIYTKMKTATNIYIFNLALADALVTTTMPFQSTVY-LMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYS----------WWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------ | |||||||||||||||||||
| 8 | 4mbsA | 0.22 | 0.23 | 0.84 | 4.28 | Download | -------------------PCQKINQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL-HYTCSSHFPYSQ----------YQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRE--KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 9 | 5o9hA | 0.28 | 0.26 | 0.87 | 3.69 | Download | ----------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYPPKVLCGVDHD--------------KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREV----LTEESVVR---- | |||||||||||||||||||
| 10 | 5c1mA | 0.26 | 0.25 | 0.85 | 4.91 | Download | -------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKY-RQGSIDCTLTFSHP-----------TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGRRITRMVLVVVAVFIVCWTPIHIYVIIKATIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
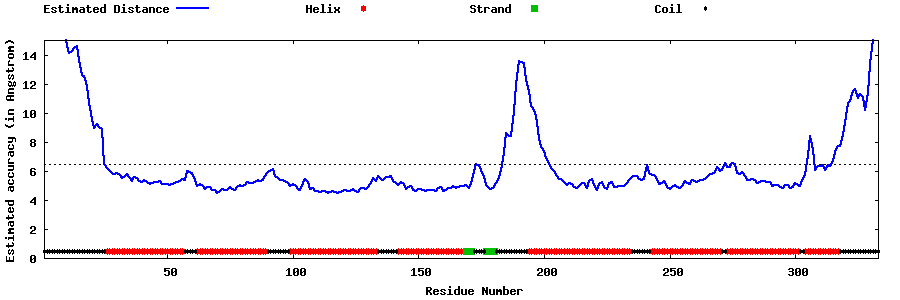
| Generated 3D models | Estimated local accuracy of models | ||
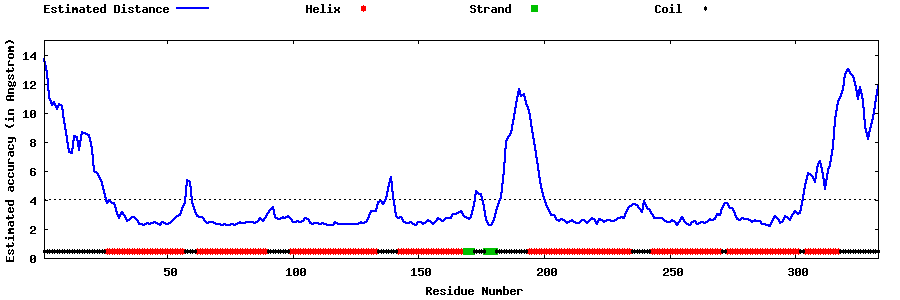 | |||
|
|
|||
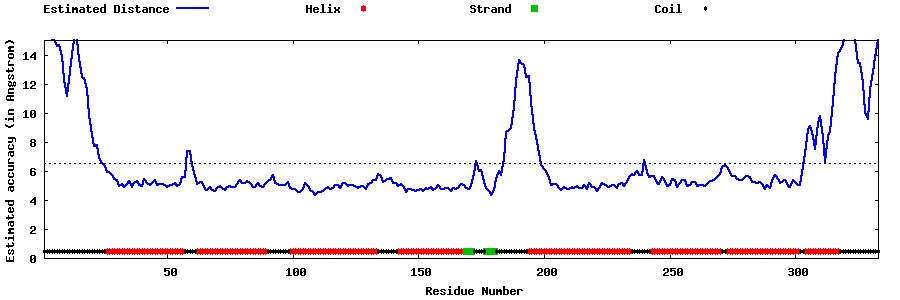 | |||
|
| |||
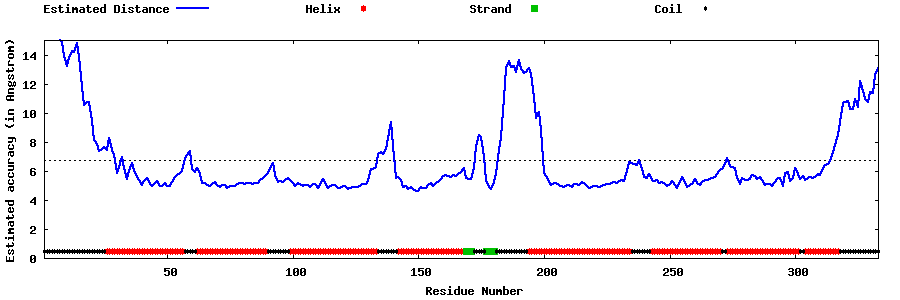 | |||
|
| |||
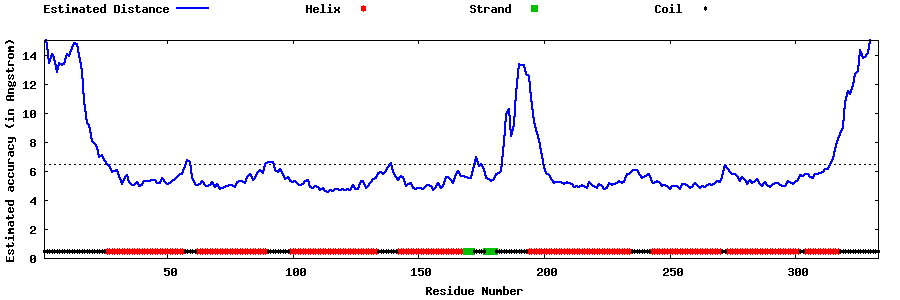 | |||
|
| |||
 | |||
|
| |||