

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGNCLHRAELSPSTENSSQLDFEDVWNSSYGVNDSFPDGDYGANLEAAAPCHSCNLLDDSALPFFILTSVLGILASSTVLFMLFRPLFRWQLCPGWPVLAQLAVGSALFSIVVPVLAPGLGSTRSSALCSLGYCVWYGSAFAQALLLGCHASLGHRLGAGQVPGLTLGLTVGIWGVAALLTLPVTLASGASGGLCTLIYSTELKALQATHTVACLAIFVLLPLGLFGAKGLKKALGMGPGPWMNILWAWFIFWWPHGVVLGLDFLVRSKLLLLSTCLAQQALDLLLNLAEALAILHCVATPLLLALFCHQATRTLLPSLPLPEGWSSHLDTLGSKS | |
| CCCCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCSSSSSSHHHHHHHHHHHCCCSSSSCCCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSSSSSSSSSSCCHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCC | |
| 987557654567767865320132036777878887567778787666777530077889999999999999998998621214437667035689999999999999999999999815775135999999999999999999999869864211222604614567437999999998564516853557751121798557999999999999999999999999999999638887675999998744251889999999997234567870699999999999999999999999999992799999999987676568889886778999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGNCLHRAELSPSTENSSQLDFEDVWNSSYGVNDSFPDGDYGANLEAAAPCHSCNLLDDSALPFFILTSVLGILASSTVLFMLFRPLFRWQLCPGWPVLAQLAVGSALFSIVVPVLAPGLGSTRSSALCSLGYCVWYGSAFAQALLLGCHASLGHRLGAGQVPGLTLGLTVGIWGVAALLTLPVTLASGASGGLCTLIYSTELKALQATHTVACLAIFVLLPLGLFGAKGLKKALGMGPGPWMNILWAWFIFWWPHGVVLGLDFLVRSKLLLLSTCLAQQALDLLLNLAEALAILHCVATPLLLALFCHQATRTLLPSLPLPEGWSSHLDTLGSKS | |
| 733223334443344333313132221332433431433323342443130443312310011211200230233122001000113333320201100000010011000000010002001001000120121113110000000000000010030232331010000000210011000000002244330102024422101101113211102221100000110231244242100000000000122001000010023141232440323300100000020200010000003101003620430130032326334434456668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCSSSSSSHHHHHHHHHHHCCCSSSSCCCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSSSSSSSSSSCCHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCC MGNCLHRAELSPSTENSSQLDFEDVWNSSYGVNDSFPDGDYGANLEAAAPCHSCNLLDDSALPFFILTSVLGILASSTVLFMLFRPLFRWQLCPGWPVLAQLAVGSALFSIVVPVLAPGLGSTRSSALCSLGYCVWYGSAFAQALLLGCHASLGHRLGAGQVPGLTLGLTVGIWGVAALLTLPVTLASGASGGLCTLIYSTELKALQATHTVACLAIFVLLPLGLFGAKGLKKALGMGPGPWMNILWAWFIFWWPHGVVLGLDFLVRSKLLLLSTCLAQQALDLLLNLAEALAILHCVATPLLLALFCHQATRTLLPSLPLPEGWSSHLDTLGSKS | |||||||||||||||||||||||||
| 1 | 4xt1A | 0.19 | 0.21 | 0.83 | 2.23 | Download | ---------------------------------------DYDEDATPCVFTDVLNQSKPVTLFLYGVVFLFGSIGNFLVIFTITWRRRIQ--CSGDVYFINLAAADLLFVCTLPLWMQY---LLDSVPCTLLTACFYVAMFASLCFITEIALDRYYAIVMRYRPVACLFSIFWWIFAVIIAIPHFMVVTKKDNQCMTDYDYLEVSYPIILNVELMLGAFVIPLSVISYCYYRISRIRIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLKWIS-SSCEFERSLKRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLLAEFR------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.19 | 0.20 | 0.80 | 3.48 | Download | -----------------------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLK--SMTDIYLLNLAISDLFFLLTVPFWAHYAQWDFGNTMCQLLTGLYFIGFFSGILTIDRYLAVVHAVFALKARTVGVVTSVITWVVAVFASLPNIIFTRSLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMCYSGILKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFF--GLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------- | |||||||||||||||||||
| 3 | 4xt1A | 0.17 | 0.21 | 0.83 | 3.44 | Download | ---------------------------------------DYDEDATPCVFTDVLNQSKPVTLFLYGVVFLFGSIGNFLVIFTITWRRRIQC--SGDVYFINLAAADLLFVCTLPLWMQYLLVPCTLLTACFYVAMFASLCFITEIALDRYYAIVYM--RYRPVKQACLFSIFWWIFAVIIAIPHFMVVTKKDNQCMTDYDYLEVSYPIILNVELMLGAFVIPLSVISYCYYRISRIVIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLKW-ISSSCEFERSLKRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLLAEFR------------- | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.20 | 0.75 | 1.54 | Download | -----------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMK--TATNIYIFNLALADALVTTTMPFQSTVYSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVCHP-VKLDRTPLKAKIINICIWLLSSSVGISAIVLGGTDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIILRLKSVLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------- | |||||||||||||||||||
| 5 | 5t1a | 0.21 | 0.17 | 0.77 | 1.17 | Download | ----------------------------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLK--CLTDIYLLNLAISDLLFLITLPLWAHSNEWVFGNAMCKLFTGLYHIGYFGGLLTIDRYLAIVHAVFALKTVTFGVVTSVITWLVAVFASVPGIIFTKQSVYVCGPYFPRG---WNNFHTIMRNILGLVLPLLIICYSGISRAEKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFF--GLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF----------------- | |||||||||||||||||||
| 6 | 4xt1A | 0.18 | 0.21 | 0.82 | 2.77 | Download | ----------------------------------------YDEDATPCVFTDVLNQSKPVTLFLYGVVFLFGSIGNFLVIFTITWRRRIQ--CSGDVYFINLAAADLLFVCTLPLWM---QYLLDSVPCTLLTACFYVAMFASLCFIDRYYAIVYM--RYRPVKQACLFSIFWWIFAVIIAIPHFMVVTKKDNQCMTDYDYLEVSYPIILNVELMLGAFVIPLSVICYYRISRIVARIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLKWIS-SSCEFERSLKRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLLAEFR------------- | |||||||||||||||||||
| 7 | 4ib4 | 0.12 | 0.21 | 0.75 | 1.68 | Download | -----------------------------------------------------------WAALLILMVIIPTIGGNTLVILAVSLEKKL--QYATNYFLMSLAVADLLVGLFMPIALLEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAWLISIGIAIPVPIKGETNPNNITCVL--TK-ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKVLGIVFFLFLLMWCPFFITNITLVLCD-------SCN-QTTLQMLLEIFVWIGYVSSGVNPLVYTL-FNKTFRDAFGRYITCNY------------ | |||||||||||||||||||
| 8 | 4mbsA | 0.20 | 0.18 | 0.77 | 2.75 | Download | ----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLK--SMTDIYLLNLAISDLFFLLTVPFWAHYAQWDFGNTMCQLLTGLYFIGFFSGIFFIDRYLAVVHAVFALKAR------TVITWVVAVFASLPNIIFEG----LCSSHFPYSFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREKRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGL--NNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------- | |||||||||||||||||||
| 9 | 4xt1A | 0.19 | 0.21 | 0.82 | 2.72 | Download | ---------------------------------------DYDEDATPCVFTDVLNQSKPVTLFLYGVVFLFGSIGNFLVIFTITWRRRIQ--CSGDVYFINLAAADLLFVCTLPLWMQY---LLDSVPCTLLTACFYVAMFASLCFITEIALDRYYAI-MRYRPVACLFSIFWWIFAVIIAIPHFMVVTKKDNQCMTDYDYLEVSYPIILNVELMLGAFVIPLSVISYCYYRISRIRIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLKWIS-SSCEFERSLKRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLLAEFR------------- | |||||||||||||||||||
| 10 | 4xt1A | 0.17 | 0.21 | 0.83 | 2.78 | Download | ---------------------------------------DYDEDATPCVFTDVLNQSKPVTLFLYGVVFLFGSIGNFLVIFTITWRRRIQ--CSGDVYFINLAAADLLFVCTLPLWMQYLSVPCTLLTACFYVAMFASLCFITEIALDRYYAIVYM--RYRPVKQACLFSIFWWIFAVIIAIPHFMVVTKKDNQCMTDYDYLEVSYPIILNVELMLFVIPLSVISYCYYRISRIKGRIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLKWIS-SSCEFERSLKRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLLAEFR------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
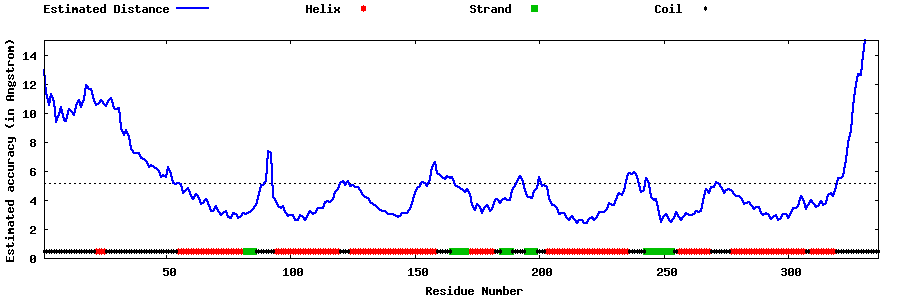 | |||
|
|
|||
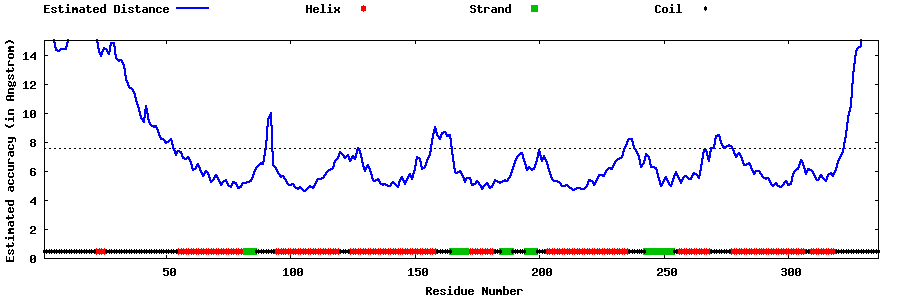 | |||
|
| |||
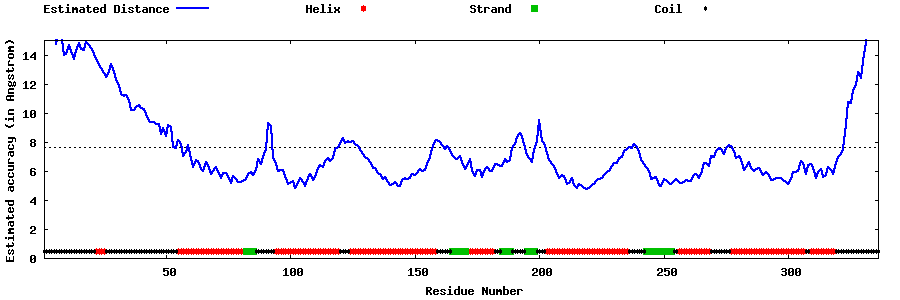 | |||
|
| |||
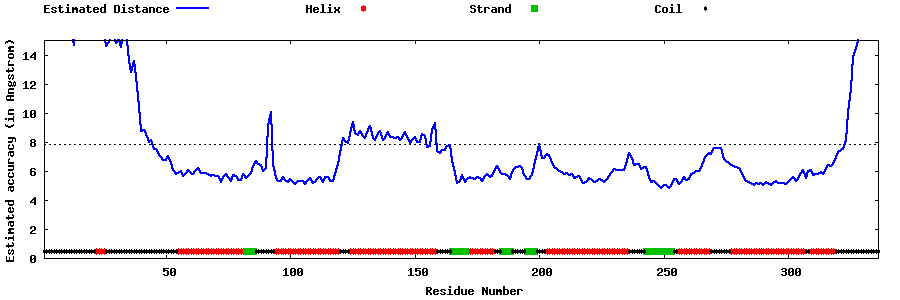 | |||
|
| |||
 | |||
|
| |||