

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MVFAHRMDNSKPHLIIPTLLVPLQNRSCTETATPLPSQYLMELSEEHSWMSNQTDLHYVLKPGEVATASIFFGILWLFSIFGNSLVCLVIHRSRRTQSTTNYFVVSMACADLLISVASTPFVLLQFTTGRWTLGSATCKVVRYFQYLTPGVQIYVLLSICIDRFYTIVYPLSFKVSREKAKKMIAASWVFDAGFVTPVLFFYGSNWDSHCNYFLPSSWEGTAYTVIHFLVGFVIPSVLIILFYQKVIKYIWRIGTDGRTVRRTMNIVPRTKVKTIKMFLILNLLFLLSWLPFHVAQLWHPHEQDYKKSSLVFTAITWISFSSSASKPTLYSIYNANFRRGMKETFCMSSMKCYRSNAYTITTSSRMAKKNYVGISEIPSMAKTITKDSIYDSFDREAKEKKLAWPINSNPPNTFV | |
| CCCHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCC | |
| 9511210135787787744265668888987889986210256766566678876455788589999999999999999997887713214078888544799999999999999998999999998172107458898799999999999999999999873520105777643366688843799999999999999825230144010126871133899999999999999999999999999999342698865304566888998899999999999999998999999999996337775489999999999999999999999739999999999848777787777876768775213678776536899876755887666777611110446789999998869 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MVFAHRMDNSKPHLIIPTLLVPLQNRSCTETATPLPSQYLMELSEEHSWMSNQTDLHYVLKPGEVATASIFFGILWLFSIFGNSLVCLVIHRSRRTQSTTNYFVVSMACADLLISVASTPFVLLQFTTGRWTLGSATCKVVRYFQYLTPGVQIYVLLSICIDRFYTIVYPLSFKVSREKAKKMIAASWVFDAGFVTPVLFFYGSNWDSHCNYFLPSSWEGTAYTVIHFLVGFVIPSVLIILFYQKVIKYIWRIGTDGRTVRRTMNIVPRTKVKTIKMFLILNLLFLLSWLPFHVAQLWHPHEQDYKKSSLVFTAITWISFSSSASKPTLYSIYNANFRRGMKETFCMSSMKCYRSNAYTITTSSRMAKKNYVGISEIPSMAKTITKDSIYDSFDREAKEKKLAWPINSNPPNTFV | |
| 6323330453533321121131034331341112223320332344433332333232312200100102211200210130010000000023402000000020001001010000010100010033011010000000000000000000000000000010000002132133100000000001000000000000204332201010126303200000001210330010002000000010233334354454444434422100000000000000000211000000101144241010000000100020000000000000630040013002013232344443434444444444433344343444333443335534463554435353564345324 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCC MVFAHRMDNSKPHLIIPTLLVPLQNRSCTETATPLPSQYLMELSEEHSWMSNQTDLHYVLKPGEVATASIFFGILWLFSIFGNSLVCLVIHRSRRTQSTTNYFVVSMACADLLISVASTPFVLLQFTTGRWTLGSATCKVVRYFQYLTPGVQIYVLLSICIDRFYTIVYPLSFKVSREKAKKMIAASWVFDAGFVTPVLFFYGSNWDSHCNYFLPSSWEGTAYTVIHFLVGFVIPSVLIILFYQKVIKYIWRIGTDGRTVRRTMNIVPRTKVKTIKMFLILNLLFLLSWLPFHVAQLWHPHEQDYKKSSLVFTAITWISFSSSASKPTLYSIYNANFRRGMKETFCMSSMKCYRSNAYTITTSSRMAKKNYVGISEIPSMAKTITKDSIYDSFDREAKEKKLAWPINSNPPNTFV | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.19 | 0.19 | 0.70 | 2.51 | Download | ----------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRAVVCMLFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.20 | 0.21 | 0.70 | 3.55 | Download | ---------------------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTDGAVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP------------------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.20 | 0.19 | 0.70 | 3.35 | Download | ----------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTDGAVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSK-----EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.20 | 0.20 | 0.84 | 1.55 | Download | -----------------------------------------DLRDNETWWYNPSIEFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYVLCNCSFDYISRDTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETE--D-DKDA-----ETEIPAGESSDAAPS------ADAAQMKE------------- | |||||||||||||||||||
| 5 | 4djh | 0.20 | 0.19 | 0.68 | 1.16 | Download | ------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNTKPNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.18 | 0.21 | 0.83 | 3.01 | Download | PEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.26 | 0.24 | 0.67 | 1.73 | Download | ----------------------------------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPRTTKMAGMMIAAAWVLSFILWAPAILFWQFIEDGECYIQFF---SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGATWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC-IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.22 | 0.17 | 0.65 | 4.10 | Download | ------------------------------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPT-----SSKAQAVNVAIWALASVVGVPVAIMQVEDEEIECLVEIPTPYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS-----REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPSSAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 4iaqA | 0.24 | 0.17 | 0.67 | 2.80 | Download | -----------------------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYKRTPKRAAVMIALVWVFSISISLPPFFWRQA---SECVVNTDH----ILYTVYSTVGAFYFPTLLLIALYGRIYVEARSR------IIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH------LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5dhgA | 0.22 | 0.17 | 0.66 | 2.63 | Download | ------------------------------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPIV--RTSSKAQAVNVAIWALASVVGVPVAIMGSEDEEIECLVEIPTPYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS-----REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
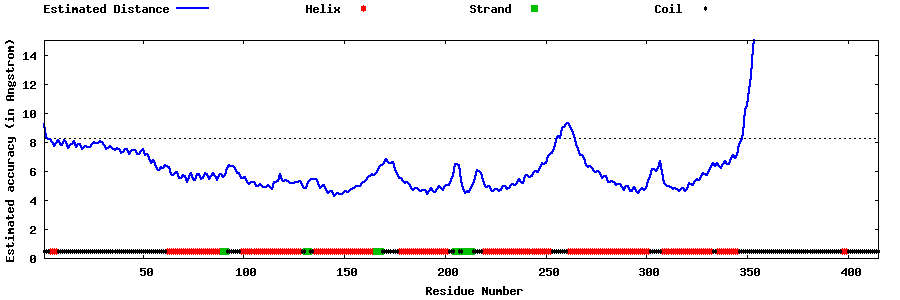 | |||
|
| |||
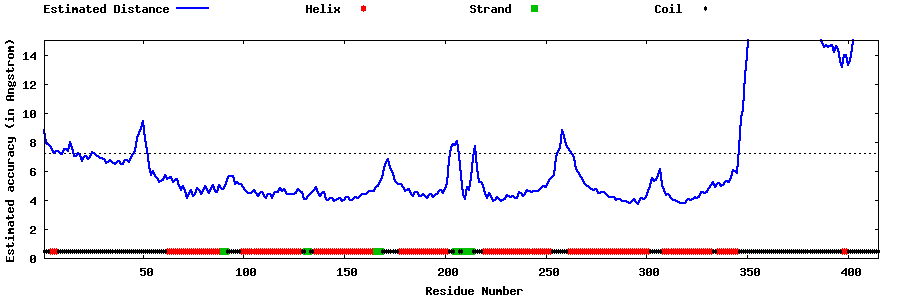 | |||
|
| |||
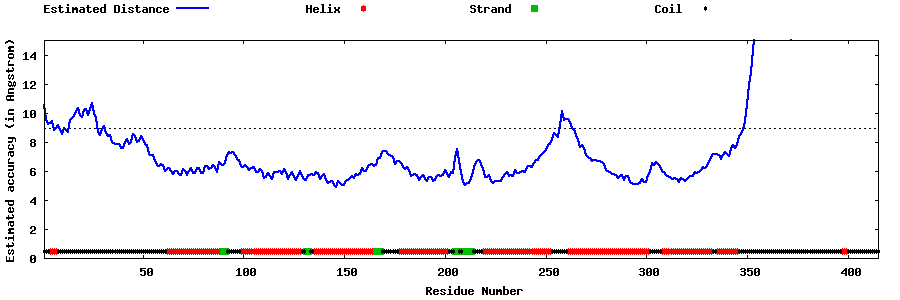 | |||
|
| |||
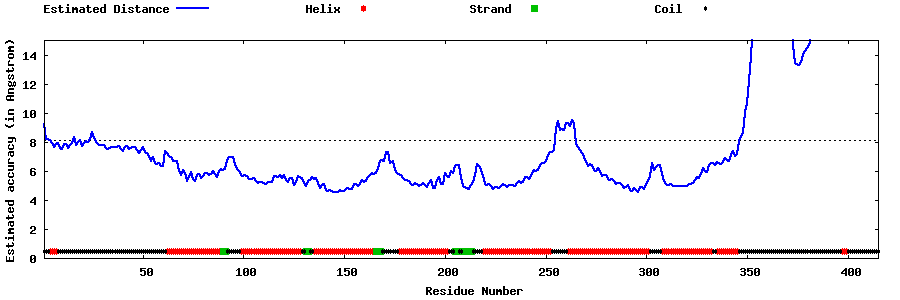 | |||
|
| |||