

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MITLNNQDQPVPFNSSHPDEYKIAALVFYSCIFIIGLFVNITALWVFSCTTKKRTTVTIYMMNVALVDLIFIMTLPFRMFYYAKDEWPFGEYFCQILGALTVFYPSIALWLLAFISADRYMAIVQPKYAKELKNTCKAVLACVGVWIMTLTTTTPLLLLYKDPDKDSTPATCLKISDIIYLKAVNVLNLTRLTFFFLIPLFIMIGCYLVIIHNLLHGRTSKLKPKVKEKSIRIIITLLVQVLVCFMPFHICFAFLMLGTGENSYNPWGAFTTFLMNLSTCLDVILYYIVSKQFQARVISVMLYRNYLRSMRRKSFRSGSLRSLSNINSEML | |
| CCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9688899999998877721899999999999999999889986684312477784869999999999999999389999999519998888788899999999999999999999998168886321011055604576677999999999998998830127759981799604994018899999999999999999999999999999998257776433100002234567899999998362989999999955107799999999999999999999999940899999999996636766544677888998778888777789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MITLNNQDQPVPFNSSHPDEYKIAALVFYSCIFIIGLFVNITALWVFSCTTKKRTTVTIYMMNVALVDLIFIMTLPFRMFYYAKDEWPFGEYFCQILGALTVFYPSIALWLLAFISADRYMAIVQPKYAKELKNTCKAVLACVGVWIMTLTTTTPLLLLYKDPDKDSTPATCLKISDIIYLKAVNVLNLTRLTFFFLIPLFIMIGCYLVIIHNLLHGRTSKLKPKVKEKSIRIIITLLVQVLVCFMPFHICFAFLMLGTGENSYNPWGAFTTFLMNLSTCLDVILYYIVSKQFQARVISVMLYRNYLRSMRRKSFRSGSLRSLSNINSEML | |
| 7444335444243434344011000021012002301321220010001223432000000000010010000000010000036411003001200121123011100200000031001000100202422323100000010010010001010001203447622100000246532312100111111301331221002001000110241444654454220000000000000100122101000010003334001001200100013000110202000044015101400323422443445433444443444443644 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MITLNNQDQPVPFNSSHPDEYKIAALVFYSCIFIIGLFVNITALWVFSCTTKKRTTVTIYMMNVALVDLIFIMTLPFRMFYYAKDEWPFGEYFCQILGALTVFYPSIALWLLAFISADRYMAIVQPKYAKELKNTCKAVLACVGVWIMTLTTTTPLLLLYKDPDKDSTPATCLKISDIIYLKAVNVLNLTRLTFFFLIPLFIMIGCYLVIIHNLLHGRTSKLKPKVKEKSIRIIITLLVQVLVCFMPFHICFAFLMLGTGENSYNPWGAFTTFLMNLSTCLDVILYYIVSKQFQARVISVMLYRNYLRSMRRKSFRSGSLRSLSNINSEML | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.22 | 0.23 | 0.89 | 3.37 | Download | ---------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP--------------------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.24 | 0.24 | 0.88 | 4.36 | Download | --------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKEEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR-------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.23 | 0.89 | 3.96 | Download | ---------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSP-SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.24 | 0.24 | 0.85 | 1.55 | Download | -----------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.24 | 0.24 | 0.85 | 1.17 | Download | -----------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA---LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.23 | 0.89 | 3.48 | Download | -----------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.24 | 0.24 | 0.85 | 1.72 | Download | ------------------PAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF----------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.22 | 0.23 | 0.90 | 4.58 | Download | --------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK-----PCG-------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.21 | 0.23 | 0.87 | 3.48 | Download | ----------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA-AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE---KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.23 | 0.24 | 0.92 | 4.05 | Download | LKTTRNAYGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
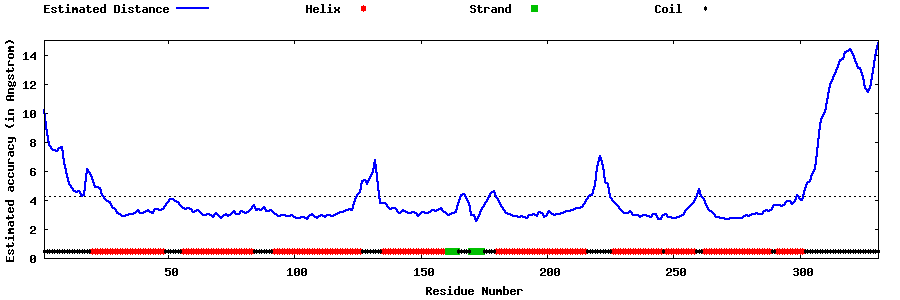
| Generated 3D models | Estimated local accuracy of models | ||
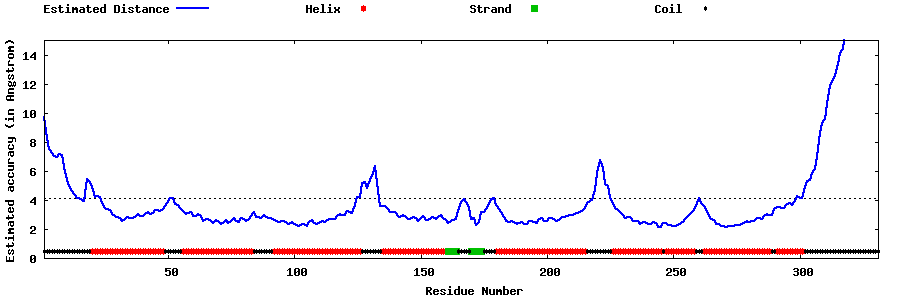 | |||
|
|
|||
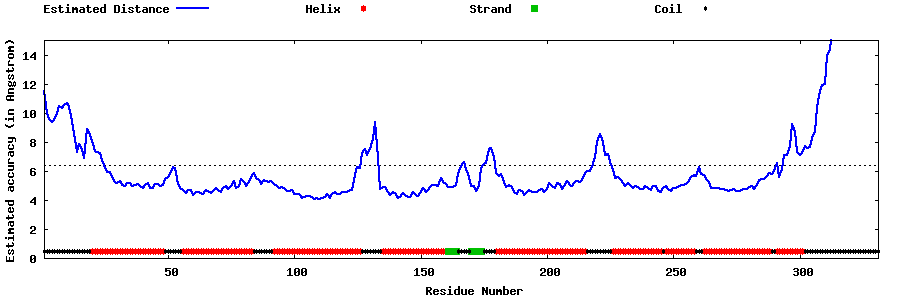 | |||
|
| |||
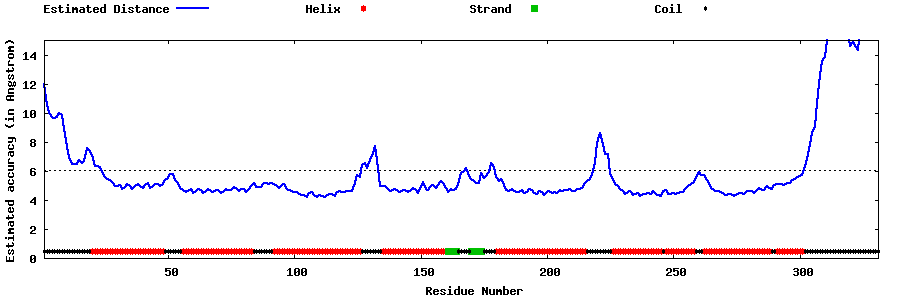 | |||
|
| |||
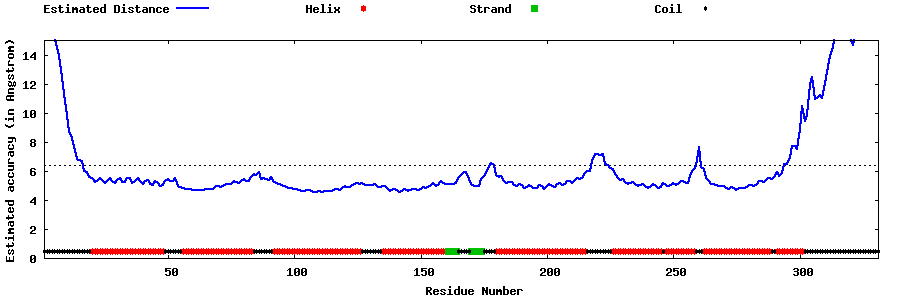 | |||
|
| |||
 | |||
|
| |||