

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MKSPFYRCQNTTSVEKGNSAVMGGVLFSTGLLGNLLALGLLARSGLGWCSRRPLRPLPSVFYMLVCGLTVTDLLGKCLLSPVVLAAYAQNRSLRVLAPALDNSLCQAFAFFMSFFGLSSTLQLLAMALECWLSLGHPFFYRRHITLRLGALVAPVVSAFSLAFCALPFMGFGKFVQYCPGTWCFIQMVHEEGSLSVLGYSVLYSSLMALLVLATVLCNLGAMRNLYAMHRRLQRHPRSCTRDCAEPRADGREASPQPLEELDHLLLLALMTVLFTMCSLPVIYRAYYGAFKDVKEKNRTSEEAEDLRALRFLSVISIVDPWIFIIFRSPVFRIFFHKIFIRPLRYRSRCSNSTNMESSL | |
| CCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCC | |
| 99999998788788866269999999999999999999988824300001321155797789999999999999999999999999982470013587677553012869999999999999999999999996471113565349899999999999999999997733203688579967985458888735334557888999889999999999999999999999998766643035555576532100667999999997878989999997489999999997587778823999999999999999999989999923999999999996699767889989998808999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MKSPFYRCQNTTSVEKGNSAVMGGVLFSTGLLGNLLALGLLARSGLGWCSRRPLRPLPSVFYMLVCGLTVTDLLGKCLLSPVVLAAYAQNRSLRVLAPALDNSLCQAFAFFMSFFGLSSTLQLLAMALECWLSLGHPFFYRRHITLRLGALVAPVVSAFSLAFCALPFMGFGKFVQYCPGTWCFIQMVHEEGSLSVLGYSVLYSSLMALLVLATVLCNLGAMRNLYAMHRRLQRHPRSCTRDCAEPRADGREASPQPLEELDHLLLLALMTVLFTMCSLPVIYRAYYGAFKDVKEKNRTSEEAEDLRALRFLSVISIVDPWIFIIFRSPVFRIFFHKIFIRPLRYRSRCSNSTNMESSL | |
| 86474141423132443110000122133133112100000001333333334344231100000000010002101111000000002222033233311100000101010000200020000000000000010131333023310000000001200100200110001122334301000002234432100000101123233312200200000000002124424434443454444434444444432330120000000001000000000000000000032322222122001120120013000110100000136003300320002113445434634445337 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCC MKSPFYRCQNTTSVEKGNSAVMGGVLFSTGLLGNLLALGLLARSGLGWCSRRPLRPLPSVFYMLVCGLTVTDLLGKCLLSPVVLAAYAQNRSLRVLAPALDNSLCQAFAFFMSFFGLSSTLQLLAMALECWLSLGHPFFYRRHITLRLGALVAPVVSAFSLAFCALPFMGFGKFVQYCPGTWCFIQMVHEEGSLSVLGYSVLYSSLMALLVLATVLCNLGAMRNLYAMHRRLQRHPRSCTRDCAEPRADGREASPQPLEELDHLLLLALMTVLFTMCSLPVIYRAYYGAFKDVKEKNRTSEEAEDLRALRFLSVISIVDPWIFIIFRSPVFRIFFHKIFIRPLRYRSRCSNSTNMESSL | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.18 | 0.20 | 0.80 | 2.20 | Download | --ENFMDIECFMVLNQLAIAVLSLTLGTFTVLENLLVLCVILHS----------RSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-------DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNC---EKLQSVCSDIFPH-----IDETYLMFWIGVTSVLLLFIVYAYMYILWKA---------------------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK----LIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------------- | |||||||||||||||||||
| 2 | 4amjA | 0.15 | 0.21 | 0.80 | 4.12 | Download | ----------------AGMSLLMALVVLLIVAGNVLVIAAIGST----------QRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTW------LWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKE------------QIRKIDRASKRKTSRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP-----DWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLL--------------------- | |||||||||||||||||||
| 3 | 2z73A | 0.11 | 0.16 | 0.92 | 2.85 | Download | VHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT----------KSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKW------IFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRD--STTRSNILCMFILFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKE------LRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIH----NPMIYSVSHPKFREAISQTFPWVLTCCQFDDDKDAETEIPA | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.21 | 0.85 | 1.53 | Download | -----------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMK----------TATNIYIFNLALADALVTT-TMPFQSTVYLMNSW-----P-FGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWWDLFMKIVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEGLTNGVTKQKQTPDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-------ALSSYYFCIALGYTNSSLNPILYAFL-DENFKRCFRDFCFP------------------ | |||||||||||||||||||
| 5 | 4ib4 | 0.15 | 0.18 | 0.86 | 1.16 | Download | ---------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLE----------KKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEA-----MWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETN---PNNITCVLTKE-----RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDAQKQKNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC--NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---------------- | |||||||||||||||||||
| 6 | 5tjvA | 0.17 | 0.20 | 0.80 | 2.62 | Download | --MDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS----------RSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-------SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNC---EKLQSVCSDIFPH-----IDETYLMFWIGVTSVLLLFIVYAYMYILWKA---------------------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK----LIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------------- | |||||||||||||||||||
| 7 | 4ib4 | 0.15 | 0.18 | 0.83 | 1.71 | Download | ------------------AALLILMVIIPTIGGNTLVILAVSLE----------KKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFE-----AMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIGIETN---PNNITCVLTK----E-RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNRAQKQLNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC--NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNY----------------- | |||||||||||||||||||
| 8 | 1u19A | 0.16 | 0.20 | 0.89 | 4.28 | Download | VRSPFEAPQYYLAWQFSMLAAYMFLLIMLGFPINFLTLYVTVQH----------KKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGY------FVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPM-SNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPEETNNESFVIYMFVVHFIIPLIVIFFCYG------------------QLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFG----PIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTTETSQV | |||||||||||||||||||
| 9 | 4n6hA | 0.12 | 0.16 | 0.84 | 2.13 | Download | --SPG-ARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY----------TKMKTATNIYIFNLALADALATSTLPFQSAKYLMET-------WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMA-VTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV-------------------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------- | |||||||||||||||||||
| 10 | 4ww3A | 0.12 | 0.16 | 0.92 | 2.34 | Download | -ETWWYNPSIVVHVPDAVLGIFIGICGIIGCGGNGIVIYLFTKT----------KSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWI------FGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYI--SRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKEL-----RKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAK----ASAIHNPMIYSVSHPKFREAISQTVLTCCQFDDKETEDDKDAETEI | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
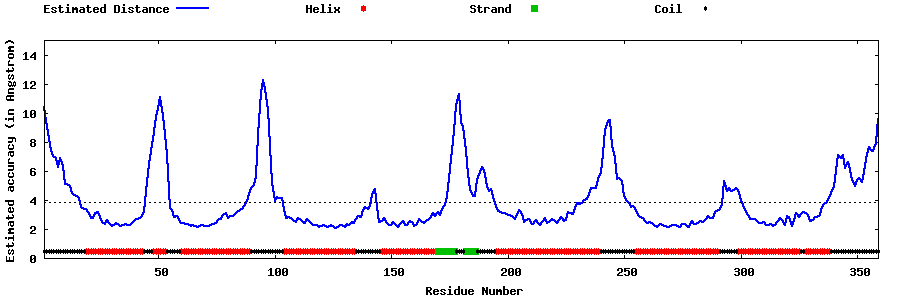 | |||
|
|
|||
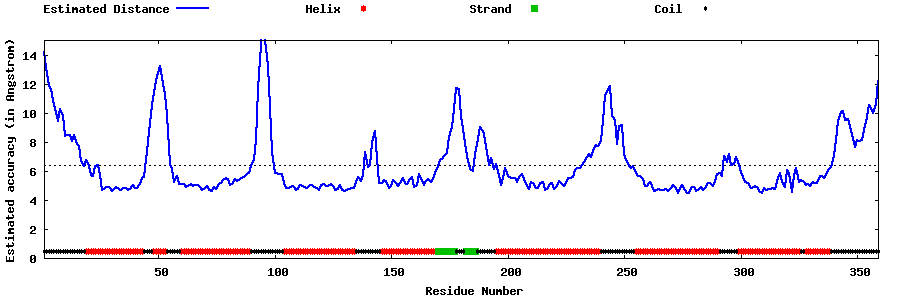 | |||
|
| |||
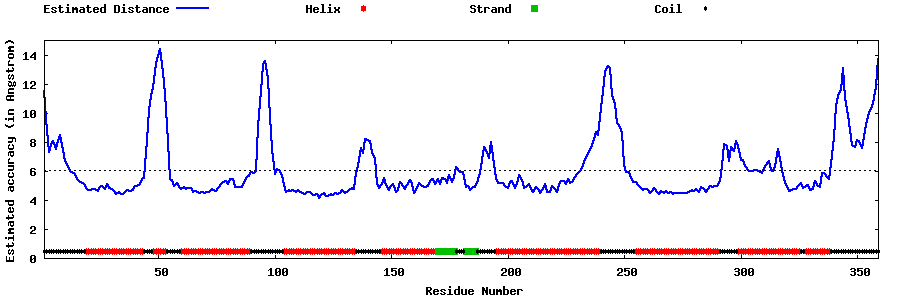 | |||
|
| |||
 | |||
|
| |||
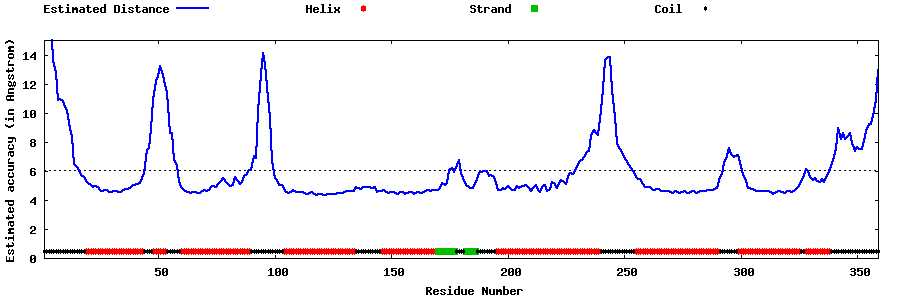 | |||
|
| |||