

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MATVNTDATDKDISKFKVTFTLVVSGIECITGILGSGFITAIYGAEWARGKTLPTGDRIMLMLSFSRLLLQIWMMLENIFSLLFRIVYNQNSVYILFKVITVFLNHSNLWFAAWLKVFYCLRIANFNHPLFFLMKRKIIVLMPWLLRLSVLVSLSFSFPLSRDVFNVYVNSSIPIPSSNSTEKKYFSETNMVNLVFFYNMGIFVPLIMFILAATLLILSLKRHTLHMGSNATGSRDPSMKAHIGAIKATSYFLILYIFNAIALFLSTSNIFDTYSSWNILCKIIMAAYPAGHSVQLILGNPGLRRAWKRFQHQVPLYLKGQTL | |
| CCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCCCCCCCC | |
| 98667899877760079999999999999999999999999999999946978878899999999999999999998794302654245644799999999998299999999999998546408998299999986221189999999999999999999876201134312357881277303421689999999999999999999999999999999999997477999999971899999999999999999999999999998727775999999999875497888999608719999999999967343277889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MATVNTDATDKDISKFKVTFTLVVSGIECITGILGSGFITAIYGAEWARGKTLPTGDRIMLMLSFSRLLLQIWMMLENIFSLLFRIVYNQNSVYILFKVITVFLNHSNLWFAAWLKVFYCLRIANFNHPLFFLMKRKIIVLMPWLLRLSVLVSLSFSFPLSRDVFNVYVNSSIPIPSSNSTEKKYFSETNMVNLVFFYNMGIFVPLIMFILAATLLILSLKRHTLHMGSNATGSRDPSMKAHIGAIKATSYFLILYIFNAIALFLSTSNIFDTYSSWNILCKIIMAAYPAGHSVQLILGNPGLRRAWKRFQHQVPLYLKGQTL | |
| 73433454363403302111112133312331332122002101110044330310010000001010000001233100000003113432000000011133123001000000000000002031100010133032100110131223222101111132233333333433421111223333220011111333333323331332332022002202330342344341131400220020002023223311211331011203442201011103223310000001304142013001300220202146474 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCCCCCCCC MATVNTDATDKDISKFKVTFTLVVSGIECITGILGSGFITAIYGAEWARGKTLPTGDRIMLMLSFSRLLLQIWMMLENIFSLLFRIVYNQNSVYILFKVITVFLNHSNLWFAAWLKVFYCLRIANFNHPLFFLMKRKIIVLMPWLLRLSVLVSLSFSFPLSRDVFNVYVNSSIPIPSSNSTEKKYFSETNMVNLVFFYNMGIFVPLIMFILAATLLILSLKRHTLHMGSNATGSRDPSMKAHIGAIKATSYFLILYIFNAIALFLSTSNIFDTYSSWNILCKIIMAAYPAGHSVQLILGNPGLRRAWKRFQHQVPLYLKGQTL | |||||||||||||||||||||||||
| 1 | 4grvA | 0.13 | 0.18 | 0.88 | 1.34 | Download | -----NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA----RKSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAIC---HPFKAK--LMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVM--------SGSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQWHYFYMLTNALAYASSAINPILYNLVSANFRQV----------------- | |||||||||||||||||||
| 2 | 5tjvA | 0.10 | 0.20 | 0.88 | 2.72 | Download | -ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS--RSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRN-VFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPK----AVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHID----------ETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------ | |||||||||||||||||||
| 3 | 4xt1A | 0.12 | 0.20 | 0.87 | 2.22 | Download | YDEDATPCVFTDVLNQSKPVTLFLYGVVFLFGSIGNFLVIFTITW---RRRIQCSGDVYFINLAAADLLFVC------TLPLWMQYLLDSVP-CTLLTACFYVAMFASLCFITEIALDRYYAIVYMRYRP--------VKQACLFSIFWWIFAVIIAIPHFMVV-TKKDNQCMTDYDY------LEVSYPIILNVELMLGAFVIPLSVISYCYYRISRIVAV--------SQSRH--KGRIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLKWIKRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLLAEFR-------- | |||||||||||||||||||
| 4 | 4djh | 0.12 | 0.19 | 0.90 | 1.56 | Download | -------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY---TKMKTATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAV---CHPVKAL-DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVECSLQFPDDDY-SWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFTPAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------- | |||||||||||||||||||
| 5 | 4grv | 0.11 | 0.16 | 0.90 | 1.24 | Download | -----NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARK----SLQSTVHYHLGSLALSDLLILLLAMPVELYNFIHHPWAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAICHPFKAK----TLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADG-THPGGLVCTPIVDTTVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMVNILRITWDAYGSGSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMCYISDEYFYMLTNALAYASSAINPILYNLVSANFRQV----------------- | |||||||||||||||||||
| 6 | 4grvA | 0.12 | 0.18 | 0.87 | 1.50 | Download | --------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA----RKSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAICH----PFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVM--------SGSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQWHYFYMLTNALAYASSAINPILYNLVSANFRQV----------------- | |||||||||||||||||||
| 7 | 3uon | 0.10 | 0.18 | 0.88 | 1.72 | Download | -----------------VVFIVLVAGSLSLVTIIGNILVMVSIK---VNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVE-D-GE-CYI----QFFSNA--AVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIGAWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------- | |||||||||||||||||||
| 8 | 4n6hA | 0.10 | 0.19 | 0.91 | 2.08 | Download | ----GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY---TKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVK----ALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTPRDGAVVCMLQFPS----PSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV-----RLLSGSDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP---CG---- | |||||||||||||||||||
| 9 | 4djhA | 0.11 | 0.19 | 0.86 | 1.42 | Download | -------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL------------DRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSASSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------- | |||||||||||||||||||
| 10 | 5c1mA | 0.11 | 0.18 | 0.89 | 1.44 | Download | --GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRY---TKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAV---CHPVK-ALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCT---LTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKS-----VRMLSGSKE-KDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
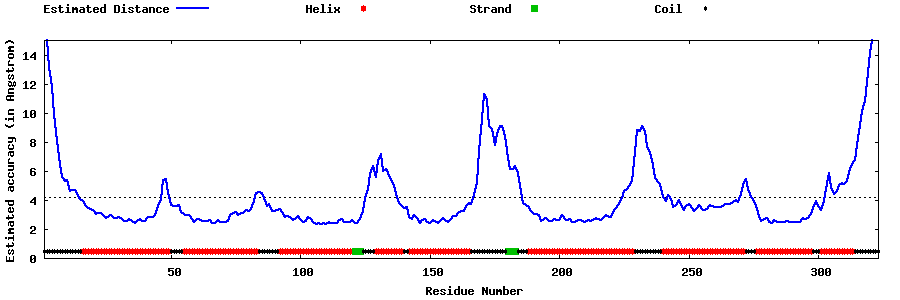
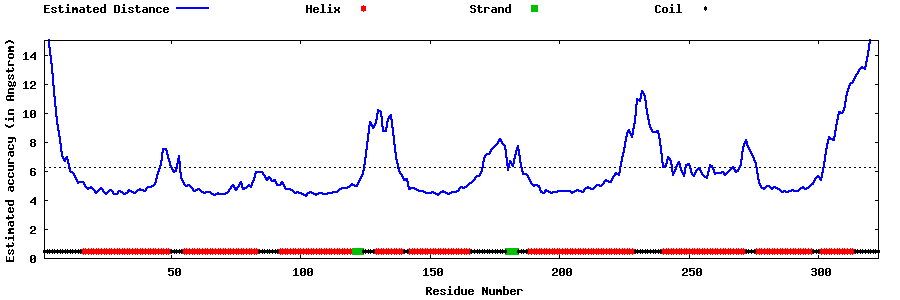
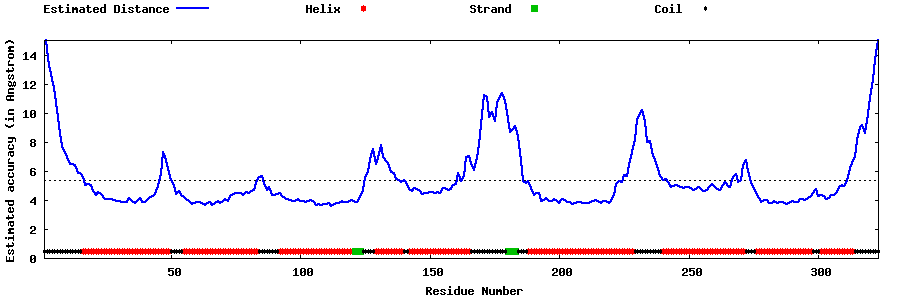
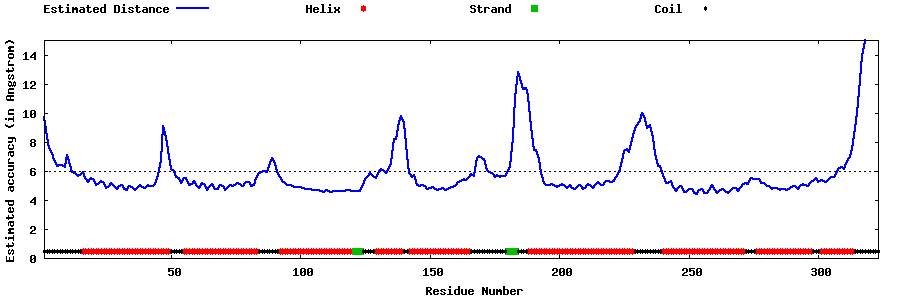
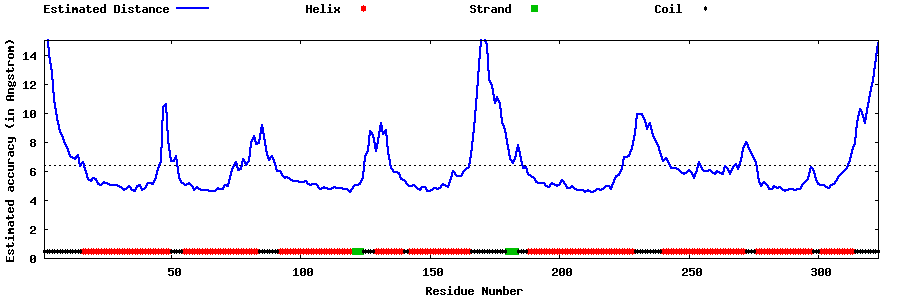
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||