

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSGESMNFSDVFDSSEDYFVSVNTSYYSVDSEMLLCSLQEVRQFSRLFVPIAYSLICVFGLLGNILVVITFAFYKKARSMTDVYLLNMAIADILFVLTLPFWAVSHATGAWVFSNATCKLLKGIYAINFNCGMLLLTCISMDRYIAIVQATKSFRLRSRTLPRSKIICLVVWGLSVIISSSTFVFNQKYNTQGSDVCEPKYQTVSEPIRWKLLMLGLELLFGFFIPLMFMIFCYTFIVKTLVQAQNSKRHKAIRVIIAVVLVFLACQIPHNMVLLVTAANLGKMNRSCQSEKLIGYTKTVTEVLAFLHCCLNPVLYAFIGQKFRNYFLKILKDLWCVRRKYKSSGFSCAGRYSENISRQTSETADNDNASSFTM | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99998886667778877677788787888876577873214878999899999999999998899989988727776749899999999999999995899999995289987758999999999999999999999999996788763275221468336759999899999999999899998235860891589512898311059999999999999999999999999999999982568877761999999999999999889999999999997367898079999999999999999999989999999408899999999974513565557788888677878888888998899998777779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSGESMNFSDVFDSSEDYFVSVNTSYYSVDSEMLLCSLQEVRQFSRLFVPIAYSLICVFGLLGNILVVITFAFYKKARSMTDVYLLNMAIADILFVLTLPFWAVSHATGAWVFSNATCKLLKGIYAINFNCGMLLLTCISMDRYIAIVQATKSFRLRSRTLPRSKIICLVVWGLSVIISSSTFVFNQKYNTQGSDVCEPKYQTVSEPIRWKLLMLGLELLFGFFIPLMFMIFCYTFIVKTLVQAQNSKRHKAIRVIIAVVLVFLACQIPHNMVLLVTAANLGKMNRSCQSEKLIGYTKTVTEVLAFLHCCLNPVLYAFIGQKFRNYFLKILKDLWCVRRKYKSSGFSCAGRYSENISRQTSETADNDNASSFTM | |
| 46444232333123342233333333133344331043630430010000210120022033232200100011343210000000000100110000000000000263010020001101001211110000000000300000000003024133310220000000002000000100000020344742310102014563321010012111110022212100020000001101315645220000000000000000122000000010023131354042230010001001000121003100000000430141014002411133463434433344434453344344344464334147 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MSGESMNFSDVFDSSEDYFVSVNTSYYSVDSEMLLCSLQEVRQFSRLFVPIAYSLICVFGLLGNILVVITFAFYKKARSMTDVYLLNMAIADILFVLTLPFWAVSHATGAWVFSNATCKLLKGIYAINFNCGMLLLTCISMDRYIAIVQATKSFRLRSRTLPRSKIICLVVWGLSVIISSSTFVFNQKYNTQGSDVCEPKYQTVSEPIRWKLLMLGLELLFGFFIPLMFMIFCYTFIVKTLVQAQNSKRHKAIRVIIAVVLVFLACQIPHNMVLLVTAANLGKMNRSCQSEKLIGYTKTVTEVLAFLHCCLNPVLYAFIGQKFRNYFLKILKDLWCVRRKYKSSGFSCAGRYSENISRQTSETADNDNASSFTM | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.34 | 0.29 | 0.79 | 3.38 | Download | ----------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKAR--TVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQ-YQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------ | |||||||||||||||||||
| 2 | 4mbsA | 0.34 | 0.29 | 0.79 | 4.27 | Download | ----------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYA-AAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTF--GVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPY-SQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.25 | 0.23 | 0.79 | 3.64 | Download | ---------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTP--AKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSP--SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-----DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.28 | 0.24 | 0.74 | 1.54 | Download | -----------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRT--PLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDD-YSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.35 | 0.29 | 0.75 | 1.18 | Download | ----------------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA-NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKART--VTFGVVTSVITWLVAVFASVPGIIFTKQKE-DSVYVCGPYFPR-----GWNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRIPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.34 | 0.29 | 0.78 | 3.36 | Download | -----------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKART--VTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPY-SQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------ | |||||||||||||||||||
| 7 | 5t1a | 0.35 | 0.29 | 0.75 | 1.70 | Download | ------------------------------------------QIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA-NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKART--VTFGVVTSVITWLVAVFASVPGIIFTKQKE-DSVYVCGPYFPR-----GWNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.35 | 0.29 | 0.78 | 5.01 | Download | ----------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKART--VTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPY-SQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK--KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------ | |||||||||||||||||||
| 9 | 4mbsA | 0.33 | 0.29 | 0.79 | 3.73 | Download | ----------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKART--VTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQY-QFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.28 | 0.25 | 0.77 | 4.22 | Download | ------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFR--TPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHP--TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKAL------ITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
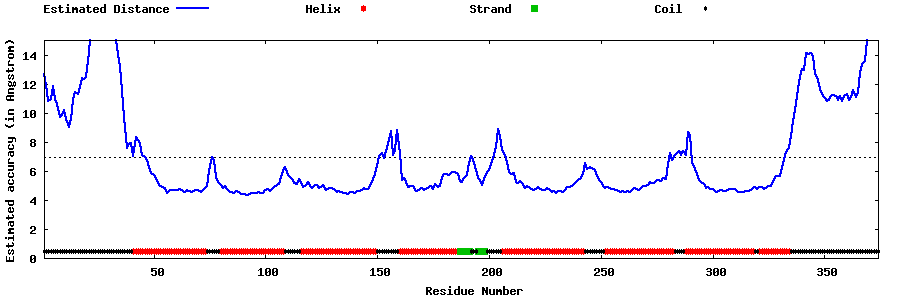
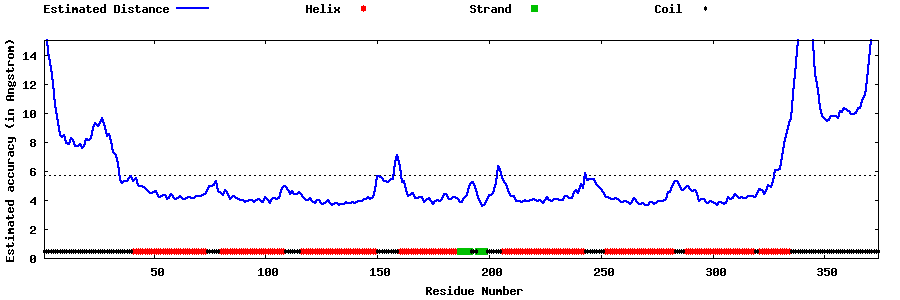
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||