

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNPTDIADTTLDESIYSNYYLYESIPKPCTKEGIKAFGELFLPPLYSLVFVFGLLGNSVVVLVLFKYKRLRSMTDVYLLNLAISDLLFVFSLPFWGYYAADQWVFGLGLCKMISWMYLVGFYSGIFFVMLMSIDRYLAIVHAVFSLRARTLTYGVITSLATWSVAVFASLPGFLFSTCYTERNHTYCKTKYSLNSTTWKVLSSLEINILGLVIPLGIMLFCYSMIIRTLQHCKNEKKNKAVKMIFAVVVLFLGFWTPYNIVLFLETLVELEVLQDCTFERYLDYAIQATETLAFVHCCLNPIIYFFLGEKFRKYILQLFKTCRGLFVLCQYCGLLQIYSADTPSSSYTQSTMDHDLHDAL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCSSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 998766667788766778777777778887434798999999999999999999889997898773667685989999999999999999899999995589967778899999999999999999999999983577552064444645234642328999999999999999703787699579961189840789999999999999999999999999999999831688776518999999999999996899999999999984688981799999999999999999999798999993388999999999976036776555677667778888888999989987303449 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNPTDIADTTLDESIYSNYYLYESIPKPCTKEGIKAFGELFLPPLYSLVFVFGLLGNSVVVLVLFKYKRLRSMTDVYLLNLAISDLLFVFSLPFWGYYAADQWVFGLGLCKMISWMYLVGFYSGIFFVMLMSIDRYLAIVHAVFSLRARTLTYGVITSLATWSVAVFASLPGFLFSTCYTERNHTYCKTKYSLNSTTWKVLSSLEINILGLVIPLGIMLFCYSMIIRTLQHCKNEKKNKAVKMIFAVVVLFLGFWTPYNIVLFLETLVELEVLQDCTFERYLDYAIQATETLAFVHCCLNPIIYFFLGEKFRKYILQLFKTCRGLFVLCQYCGLLQIYSADTPSSSYTQSTMDHDLHDAL | |
| 744643423333343333323344343204363043001000021012002302321220010001133321000000000010011000000000000032010020001201001210011000000000300000000003024223221010000000100000010000002034474221010202473220100111121121222122000200010011023146453200000000000001003110100000100231412440323200100010010001210131000000004401410140024133343446334334444454444443443444433647 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCSSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MNPTDIADTTLDESIYSNYYLYESIPKPCTKEGIKAFGELFLPPLYSLVFVFGLLGNSVVVLVLFKYKRLRSMTDVYLLNLAISDLLFVFSLPFWGYYAADQWVFGLGLCKMISWMYLVGFYSGIFFVMLMSIDRYLAIVHAVFSLRARTLTYGVITSLATWSVAVFASLPGFLFSTCYTERNHTYCKTKYSLNSTTWKVLSSLEINILGLVIPLGIMLFCYSMIIRTLQHCKNEKKNKAVKMIFAVVVLFLGFWTPYNIVLFLETLVELEVLQDCTFERYLDYAIQATETLAFVHCCLNPIIYFFLGEKFRKYILQLFKTCRGLFVLCQYCGLLQIYSADTPSSSYTQSTMDHDLHDAL | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.52 | 0.43 | 0.81 | 3.48 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.52 | 0.42 | 0.81 | 4.45 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.52 | 0.43 | 0.81 | 3.94 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.26 | 0.24 | 0.77 | 1.55 | Download | ----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-------A----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.51 | 0.41 | 0.78 | 1.18 | Download | ---------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAANEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQK-EDSVYVCGPYFPRG---WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRIPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF----------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.52 | 0.43 | 0.81 | 3.46 | Download | ----------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------------- | |||||||||||||||||||
| 7 | 5t1a | 0.52 | 0.41 | 0.78 | 1.72 | Download | ----------------------------------KQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAANEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQK-EDSVYVCGPYFPRG---WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRIPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF----------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.53 | 0.42 | 0.81 | 5.37 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK--KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.53 | 0.43 | 0.81 | 3.77 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.27 | 0.24 | 0.79 | 4.44 | Download | -----------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALI------TIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
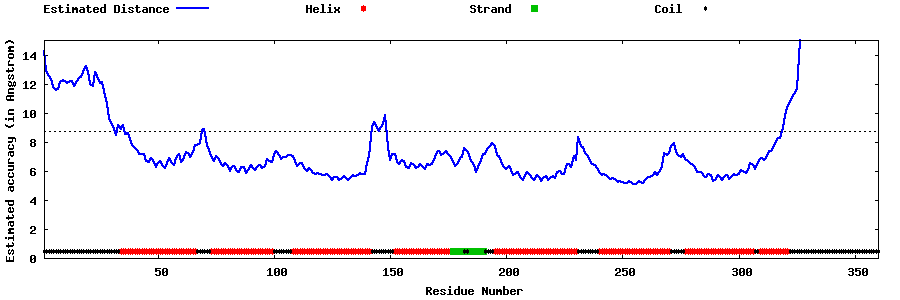
| Generated 3D models | Estimated local accuracy of models | ||
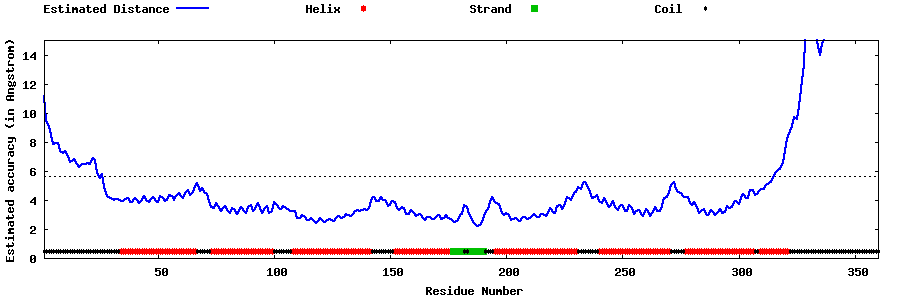 | |||
|
|
|||
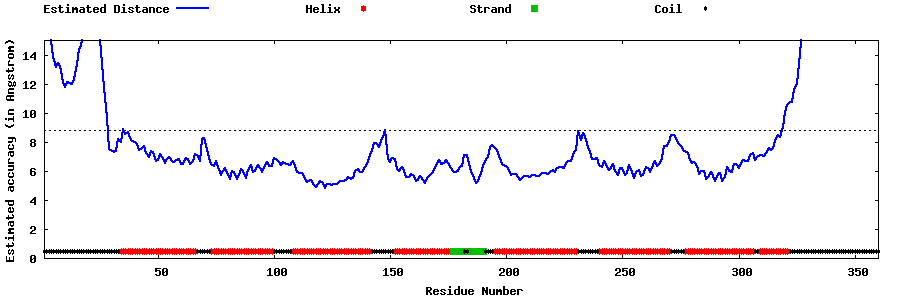 | |||
|
| |||
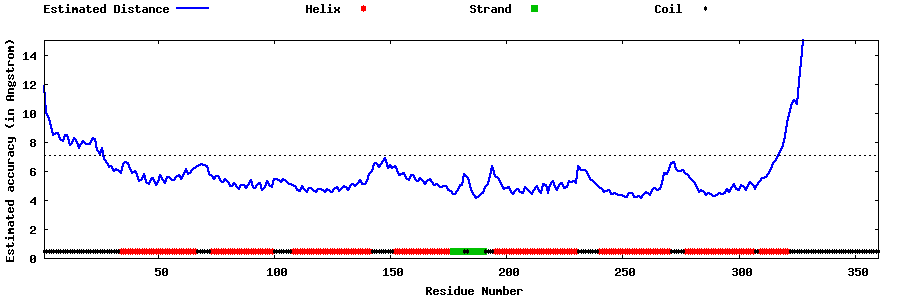 | |||
|
| |||
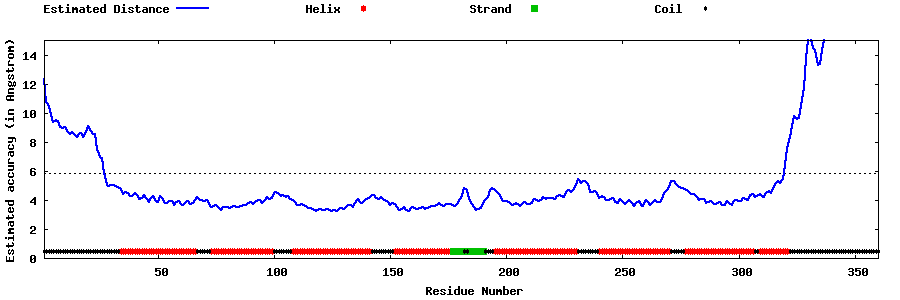 | |||
|
| |||
 | |||
|
| |||