

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MASTESSLLRSLGLSPGPGSSEVELDCWFDEDFKFILLPVSYAVVFVLGLGLNAPTLWLFIFRLRPWDATATYMFHLALSDTLYVLSLPTLIYYYAAHNHWPFGTEICKFVRFLFYWNLYCSVLFLTCISVHRYLGICHPLRALRWGRPRLAGLLCLAVWLVVAGCLVPNLFFVTTSNKGTTVLCHDTTRPEEFDHYVHFSSAVMGLLFGVPCLVTLVCYGLMARRLYQPLPGSAQSSSRLRSLRTIAVVLTVFAVCFVPFHITRTIYYLARLLEADCRVLNIVNVVYKVTRPLASANSCLDPVLYLLTGDKYRRQLRQLCGGGKPQPRTAASSLALVSLPEDSSCRWAATPQDSSCSTPRADRL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99988754357788999999887877776611889989999999999999989997885540478886889999999999999999749999999568997788678999999999999999999999999705776420422236561125555699999999999899881003735994699467980467899999999999999999999999999999997575676444321101578888989999998468999999999998647991699999999999999999999998999993488999999998540367876777876656678888888888988889999986779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MASTESSLLRSLGLSPGPGSSEVELDCWFDEDFKFILLPVSYAVVFVLGLGLNAPTLWLFIFRLRPWDATATYMFHLALSDTLYVLSLPTLIYYYAAHNHWPFGTEICKFVRFLFYWNLYCSVLFLTCISVHRYLGICHPLRALRWGRPRLAGLLCLAVWLVVAGCLVPNLFFVTTSNKGTTVLCHDTTRPEEFDHYVHFSSAVMGLLFGVPCLVTLVCYGLMARRLYQPLPGSAQSSSRLRSLRTIAVVLTVFAVCFVPFHITRTIYYLARLLEADCRVLNIVNVVYKVTRPLASANSCLDPVLYLLTGDKYRRQLRQLCGGGKPQPRTAASSLALVSLPEDSSCRWAATPQDSSCSTPRADRL | |
| 74444343243332333333343434142445021000022012002302311010000001223432101000000000000000002010000023443200400111002111311100010000003100000010020242333210000000010000000101000120355732210102034731311101111111311331120002000100110133356346443110000000000000010123101000010013023340413310200110010000300021020000004401410240034244464444444344434444344445445545354553646 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MASTESSLLRSLGLSPGPGSSEVELDCWFDEDFKFILLPVSYAVVFVLGLGLNAPTLWLFIFRLRPWDATATYMFHLALSDTLYVLSLPTLIYYYAAHNHWPFGTEICKFVRFLFYWNLYCSVLFLTCISVHRYLGICHPLRALRWGRPRLAGLLCLAVWLVVAGCLVPNLFFVTTSNKGTTVLCHDTTRPEEFDHYVHFSSAVMGLLFGVPCLVTLVCYGLMARRLYQPLPGSAQSSSRLRSLRTIAVVLTVFAVCFVPFHITRTIYYLARLLEADCRVLNIVNVVYKVTRPLASANSCLDPVLYLLTGDKYRRQLRQLCGGGKPQPRTAASSLALVSLPEDSSCRWAATPQDSSCSTPRADRL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.29 | 0.28 | 0.88 | 2.94 | Download | EAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.41 | 0.36 | 0.82 | 4.30 | Download | ---------------------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKEEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLTPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR---------------------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.22 | 0.20 | 0.81 | 3.72 | Download | -----------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRM---KEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.29 | 0.25 | 0.77 | 1.54 | Download | -----------------------------SP-AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS----A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------ | |||||||||||||||||||
| 5 | 4xnv | 0.41 | 0.35 | 0.80 | 1.17 | Download | ---------------------SFKCA-LTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYTEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.22 | 0.20 | 0.80 | 3.34 | Download | ------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEE---EKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.29 | 0.25 | 0.77 | 1.68 | Download | --------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVY-LMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------ | |||||||||||||||||||
| 8 | 4mbsA | 0.22 | 0.20 | 0.79 | 4.83 | Download | -----------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK-------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.22 | 0.20 | 0.81 | 3.51 | Download | -----------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK---EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.30 | 0.26 | 0.80 | 4.38 | Download | -------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-----TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
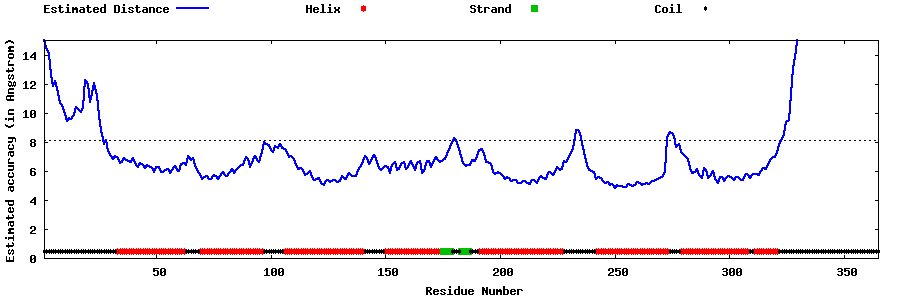
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||