

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDPEETSVYLDYYYATSPNSDIRETHSHVPYTSVFLPVFYTAVFLTGVLGNLVLMGALHFKPGSRRLIDIFIINLAASDFIFLVTLPLWVDKEASLGLWRTGSFLCKGSSYMISVNMHCSVLLLTCMSVDRYLAIVWPVVSRKFRRTDCAYVVCASIWFISCLLGLPTLLSRELTLIDDKPYCAEKKATPIKLIWSLVALIFTFFVPLLSIVTCYCCIARKLCAHYQQSGKHNKKLKKSIKIIFIVVAAFLVSWLPFNTFKFLAIVSGLRQEHYLPSAILQLGMEVSGPLAFANSCVNPFIYYIFDSYIRRAIVHCLCPCLKNYDFGSSTETSDSHLTKALSTFIHAEDFARRRKRSVSL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 997557887766678888877443333212899969999999999999789986684421777686999999999999999999819999999668988687768989999999999999999999999847887630620126351657555699999999999899998337853993687188981799999999999999999999999999999998242467510001001288999999999999857999999999998744367418899999999999999999969899999728889999999977772688878777778888566866763688766678888889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDPEETSVYLDYYYATSPNSDIRETHSHVPYTSVFLPVFYTAVFLTGVLGNLVLMGALHFKPGSRRLIDIFIINLAASDFIFLVTLPLWVDKEASLGLWRTGSFLCKGSSYMISVNMHCSVLLLTCMSVDRYLAIVWPVVSRKFRRTDCAYVVCASIWFISCLLGLPTLLSRELTLIDDKPYCAEKKATPIKLIWSLVALIFTFFVPLLSIVTCYCCIARKLCAHYQQSGKHNKKLKKSIKIIFIVVAAFLVSWLPFNTFKFLAIVSGLRQEHYLPSAILQLGMEVSGPLAFANSCVNPFIYYIFDSYIRRAIVHCLCPCLKNYDFGSSTETSDSHLTKALSTFIHAEDFARRRKRSVSL | |
| 823744332211313326434214444234001000021012002303323320110001233321000000000010110000000010000033642100200122010113211110000000002000000000020242232110000000011000000101000020353742210103115401100111111302221112022300000010133345444544422000000000000010031201000001002303344132230010011001000131003100000000440141014003320643546444433444344433433347434334454436 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDPEETSVYLDYYYATSPNSDIRETHSHVPYTSVFLPVFYTAVFLTGVLGNLVLMGALHFKPGSRRLIDIFIINLAASDFIFLVTLPLWVDKEASLGLWRTGSFLCKGSSYMISVNMHCSVLLLTCMSVDRYLAIVWPVVSRKFRRTDCAYVVCASIWFISCLLGLPTLLSRELTLIDDKPYCAEKKATPIKLIWSLVALIFTFFVPLLSIVTCYCCIARKLCAHYQQSGKHNKKLKKSIKIIFIVVAAFLVSWLPFNTFKFLAIVSGLRQEHYLPSAILQLGMEVSGPLAFANSCVNPFIYYIFDSYIRRAIVHCLCPCLKNYDFGSSTETSDSHLTKALSTFIHAEDFARRRKRSVSL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.24 | 0.26 | 0.88 | 2.89 | Download | AAAEQLKTTRNAYIQKY-LGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.24 | 0.22 | 0.81 | 4.33 | Download | ----------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKEEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFF------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.25 | 0.82 | 3.81 | Download | --------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.26 | 0.25 | 0.78 | 1.53 | Download | ----------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.25 | 0.25 | 0.80 | 1.17 | Download | ---------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE-DSVYVCGPYFPRGWNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIFYPPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.25 | 0.25 | 0.82 | 3.29 | Download | ----------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.23 | 0.24 | 0.79 | 1.70 | Download | -----------------------------DAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTEGVLCNCSFDYISRDTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE-------WVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWV---------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.24 | 0.21 | 0.80 | 4.44 | Download | ---------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK--------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------ | |||||||||||||||||||
| 9 | 5o9hA | 0.26 | 0.23 | 0.81 | 3.44 | Download | --------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFE-AKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEPPKVLCGVDHDKRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWS------ARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFL---EPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR-------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.26 | 0.23 | 0.81 | 4.36 | Download | -----------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLM-GTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKAL-----ITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
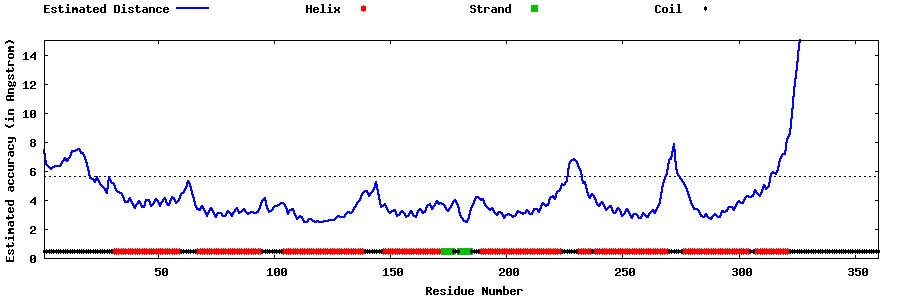 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
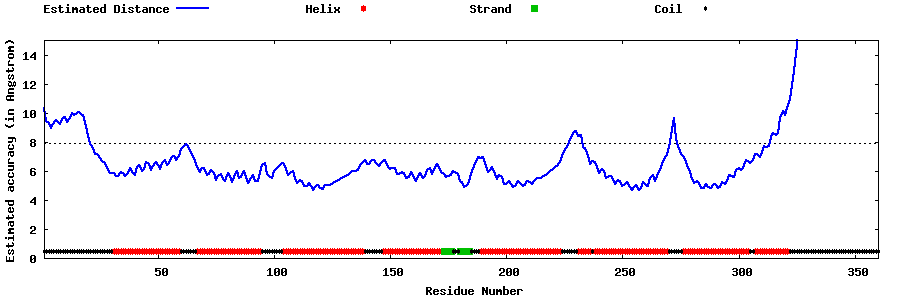 | |||
|
| |||
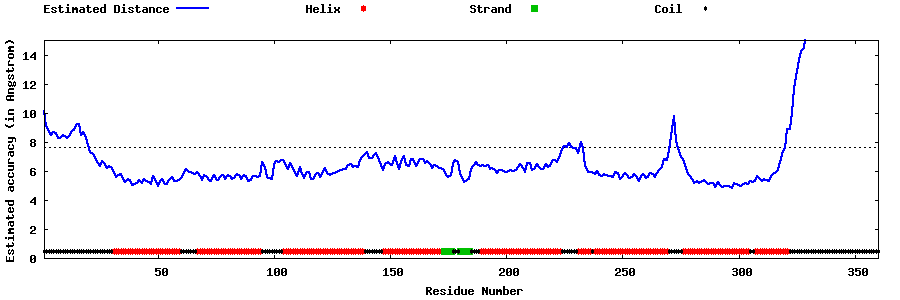 | |||
|
| |||