

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MASSTTRGPRVSDLFSGLPPAVTTPANQSAEASAGNGSVAGADAPAVTPFQSLQLVHQLKGLIVLLYSVVVVVGLVGNCLLVLVIARVRRLHNVTNFLIGNLALSDVLMCTACVPLTLAYAFEPRGWVFGGGLCHLVFFLQPVTVYVSVFTLTTIAVDRYVVLVHPLRRRISLRLSAYAVLAIWALSAVLALPAAVHTYHVELKPHDVRLCEEFWGSQERQRQLYAWGLLLVTYLLPLLVILLSYVRVSVKLRNRVVPGCVTQSQADWDRARRRRTFCLLVVIVVVFAVCWLPLHVFNLLRDLDPHAIDPYAFGLVQLLCHWLAMSSACYNPFIYAWLHDSFREELRKLLVAWPRKIAPHGQNMTVSVVI | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCC | |
| 9999999998654346899877899888777889988878888776777643353248999999999999999857889876876317989858899999999999999999625999999977996687214205999999999999999999999708783186843445637886407999999999989999867999669985899636997057999999999999999999999999999999997466988531257789888322599999999999999989999999999970032546799999999999999999999999998499999999999542369999899998730129 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MASSTTRGPRVSDLFSGLPPAVTTPANQSAEASAGNGSVAGADAPAVTPFQSLQLVHQLKGLIVLLYSVVVVVGLVGNCLLVLVIARVRRLHNVTNFLIGNLALSDVLMCTACVPLTLAYAFEPRGWVFGGGLCHLVFFLQPVTVYVSVFTLTTIAVDRYVVLVHPLRRRISLRLSAYAVLAIWALSAVLALPAAVHTYHVELKPHDVRLCEEFWGSQERQRQLYAWGLLLVTYLLPLLVILLSYVRVSVKLRNRVVPGCVTQSQADWDRARRRRTFCLLVVIVVVFAVCWLPLHVFNLLRDLDPHAIDPYAFGLVQLLCHWLAMSSACYNPFIYAWLHDSFREELRKLLVAWPRKIAPHGQNMTVSVVI | |
| 8635435424133314323232333333123333332323333333334243231220000000211120020023132000000001340200000002000302111010011010000001431000200110010010000000000000000000200000023322331000000001100000010000001013145441100101013662021000000032112310100020001000102334332544544444334421000000000000000002220000001001222243200100101000000000000000100004300400240021023435465532321014 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCC MASSTTRGPRVSDLFSGLPPAVTTPANQSAEASAGNGSVAGADAPAVTPFQSLQLVHQLKGLIVLLYSVVVVVGLVGNCLLVLVIARVRRLHNVTNFLIGNLALSDVLMCTACVPLTLAYAFEPRGWVFGGGLCHLVFFLQPVTVYVSVFTLTTIAVDRYVVLVHPLRRRISLRLSAYAVLAIWALSAVLALPAAVHTYHVELKPHDVRLCEEFWGSQERQRQLYAWGLLLVTYLLPLLVILLSYVRVSVKLRNRVVPGCVTQSQADWDRARRRRTFCLLVVIVVVFAVCWLPLHVFNLLRDLDPHAIDPYAFGLVQLLCHWLAMSSACYNPFIYAWLHDSFREELRKLLVAWPRKIAPHGQNMTVSVVI | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.26 | 0.29 | 0.81 | 2.80 | Download | -----------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM---TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR--DGAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------- | |||||||||||||||||||
| 2 | 5gliA | 0.23 | 0.25 | 0.83 | 3.82 | Download | --------------------------------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAED-WPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDYKGLRICLLHPVQKQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNIRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.29 | 0.81 | 3.84 | Download | -----------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.20 | 0.79 | 1.55 | Download | ------------------------------------------------EEQG-----NKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQSRATAFIKITVVWLISIGIAIPVPIKGIETN---PNNITCVLTKE----RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC-NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------- | |||||||||||||||||||
| 5 | 4djh | 0.29 | 0.27 | 0.78 | 1.16 | Download | -------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNTKPNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.27 | 0.29 | 0.80 | 3.28 | Download | -------------------------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG--AVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------- | |||||||||||||||||||
| 7 | 4ib4 | 0.19 | 0.20 | 0.78 | 1.70 | Download | ------------------------------------------------------------HWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQSRATAFIKITVVWLISIGIAIPVPIKGIETN---PNNITCVLTKE----RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC-NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.27 | 0.29 | 0.81 | 5.20 | Download | ----------------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.27 | 0.29 | 0.81 | 3.29 | Download | -----------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM---TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR--DGAVVCMLQFPSPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------- | |||||||||||||||||||
| 10 | 5dhgA | 0.31 | 0.28 | 0.76 | 3.95 | Download | -------------------------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGF-WPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPI--VRTSSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEE--IECLVEIPPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS-----REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPSS-ETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR----------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
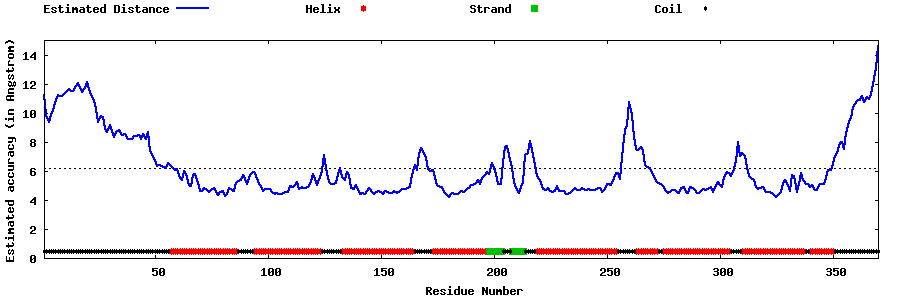
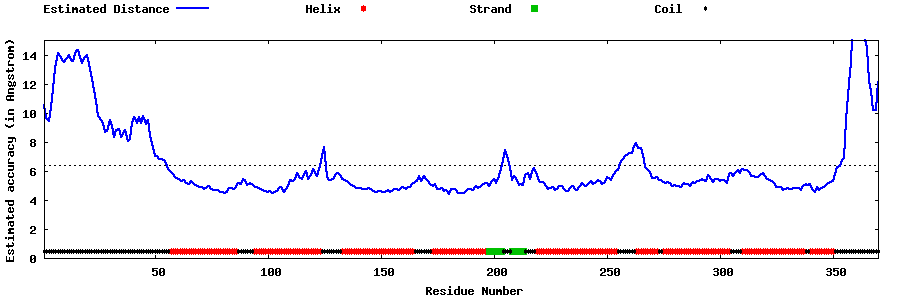
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||