

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSENGSFANCCEAGGWAVRPGWSGAGSARPSRTPRPPWVAPALSAVLIVTTAVDVVGNLLVILSVLRNRKLRNAGNLFLVSLALADLVVAFYPYPLILVAIFYDGWALGEEHCKASAFVMGLSVIGSVFNITAIAINRYCYICHSMAYHRIYRRWHTPLHICLIWLLTVVALLPNFFVGSLEYDPRIYSCTFIQTASTQYTAAVVVIHFLLPIAVVSFCYLRIWVLVLQARRKAKPESRLCLKPSDLRSFLTMFVVFVIFAICWAPLNCIGLAVAINPQEMAPQIPEGLFVTSYLLAYFNSCLNAIVYGLLNQNFRREYKRILLALWNPRHCIQDASKGSHAEGLQSPAPPIIGVQHQADAL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99987877777888887889988777888777889735999999999999999998488753103605998849999999999999999999899999999948625845898799999999999999999999997064110537443000377677510499999999999999866789469816998625762379999999989999999999999999999660576675103678898879999999999999999899999999999667422310689999999999999998999999958999999999998553678887778787788898708899988877545679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSENGSFANCCEAGGWAVRPGWSGAGSARPSRTPRPPWVAPALSAVLIVTTAVDVVGNLLVILSVLRNRKLRNAGNLFLVSLALADLVVAFYPYPLILVAIFYDGWALGEEHCKASAFVMGLSVIGSVFNITAIAINRYCYICHSMAYHRIYRRWHTPLHICLIWLLTVVALLPNFFVGSLEYDPRIYSCTFIQTASTQYTAAVVVIHFLLPIAVVSFCYLRIWVLVLQARRKAKPESRLCLKPSDLRSFLTMFVVFVIFAICWAPLNCIGLAVAINPQEMAPQIPEGLFVTSYLLAYFNSCLNAIVYGLLNQNFRREYKRILLALWNPRHCIQDASKGSHAEGLQSPAPPIIGVQHQADAL | |
| 35733233212333333233322333333343233120010000211120022023112000000001340210000002000301201010101000000026320003002100100100000000000000000001000100212331231100000000121010001110000203344310101021233200000003221331010002002100010132334254554444334420000000000001020321210000000013332432001001020010002000230001000063014001400200234443446444444454353544434434444644 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MSENGSFANCCEAGGWAVRPGWSGAGSARPSRTPRPPWVAPALSAVLIVTTAVDVVGNLLVILSVLRNRKLRNAGNLFLVSLALADLVVAFYPYPLILVAIFYDGWALGEEHCKASAFVMGLSVIGSVFNITAIAINRYCYICHSMAYHRIYRRWHTPLHICLIWLLTVVALLPNFFVGSLEYDPRIYSCTFIQTASTQYTAAVVVIHFLLPIAVVSFCYLRIWVLVLQARRKAKPESRLCLKPSDLRSFLTMFVVFVIFAICWAPLNCIGLAVAINPQEMAPQIPEGLFVTSYLLAYFNSCLNAIVYGLLNQNFRREYKRILLALWNPRHCIQDASKGSHAEGLQSPAPPIIGVQHQADAL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.22 | 0.23 | 0.82 | 3.00 | Download | ---------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.23 | 0.23 | 0.78 | 3.85 | Download | -------------------------------------ALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.23 | 0.23 | 0.82 | 3.69 | Download | ---------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.22 | 0.22 | 0.81 | 1.55 | Download | -------------------------------EEQGNK--LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIET-NPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC-NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.23 | 0.23 | 0.78 | 1.16 | Download | -----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIPYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.23 | 0.81 | 3.27 | Download | -----------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.23 | 0.23 | 0.78 | 1.70 | Download | ---------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC----IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.25 | 0.21 | 0.75 | 4.53 | Download | -----------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHP-------TSSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEEIECLVEIPTPPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS---REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQP-SSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR------------------------------------------ | |||||||||||||||||||
| 9 | 4n6hA | 0.22 | 0.23 | 0.82 | 3.08 | Download | ---------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.22 | 0.23 | 0.80 | 3.83 | Download | ------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS---KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALIT-IPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
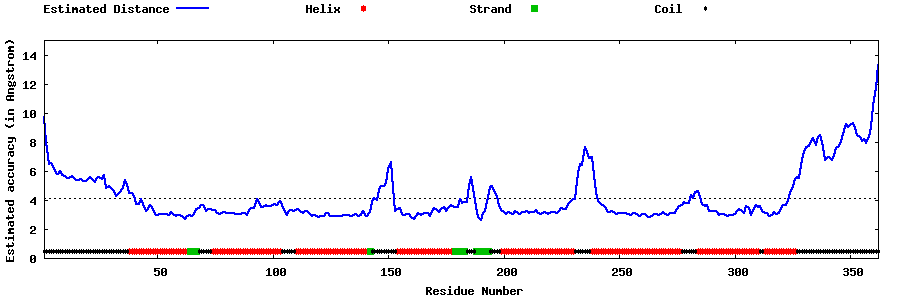
| Generated 3D models | Estimated local accuracy of models | ||
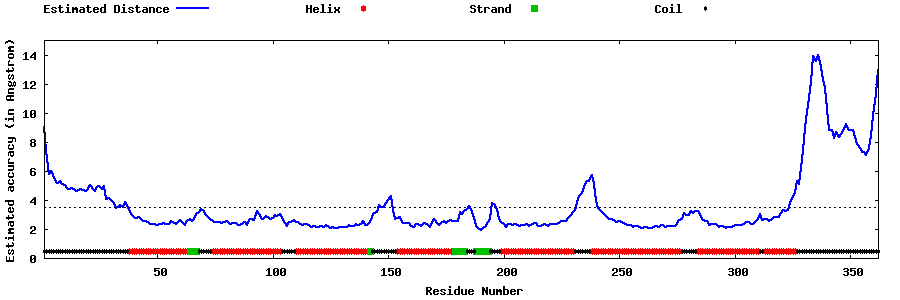 | |||
|
|
|||
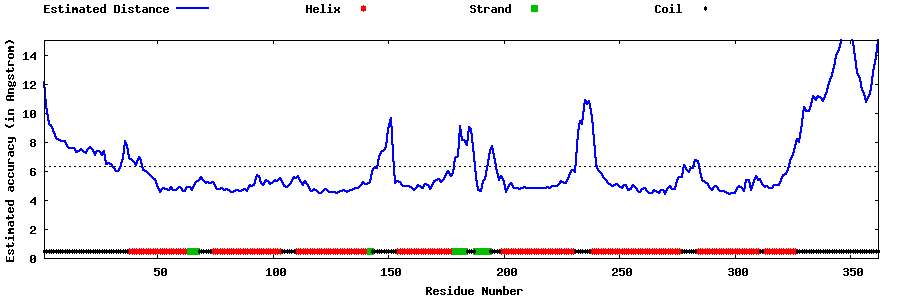 | |||
|
| |||
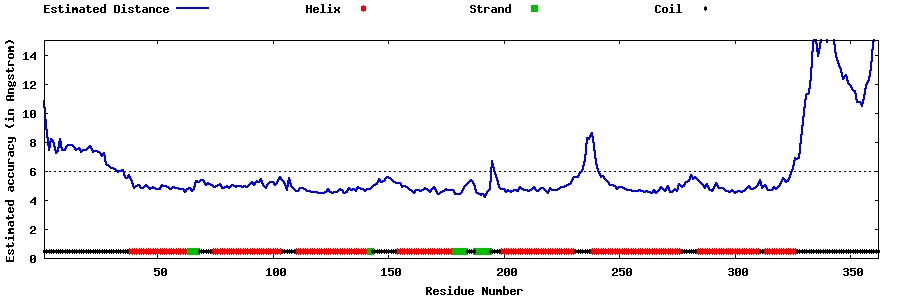 | |||
|
| |||
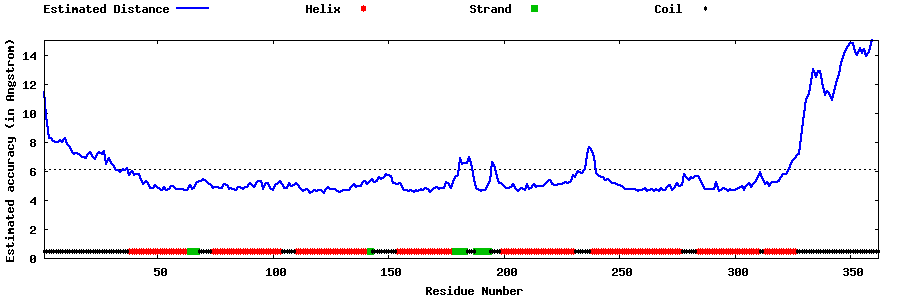 | |||
|
| |||
 | |||
|
| |||