

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDNASFSEPWPANASGPDPALSCSNASTLAPLPAPLAVAVPVVYAVICAVGLAGNSAVLYVLLRAPRMKTVTNLFILNLAIADELFTLVLPINIADFLLRQWPFGELMCKLIVAIDQYNTFSSLYFLTVMSADRYLVVLATAESRRVAGRTYSAARAVSLAVWGIVTLVVLPFAVFARLDDEQGRRQCVLVFPQPEAFWWRASRLYTLVLGFAIPVSTICVLYTTLLCRLHAMRLDSHAKALERAKKRVTFLVVAILAVCLLCWTPYHLSTVVALTTDLPQTPLVIAISYFITSLSYANSCLNPFLYAFLDASFRRNLRQLITCRAAA | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCC | |
| 9987689988887788887657677654666422899859999999999988779984575631998785999999999999999999579999999608998966688899999999999999999999997060132117645554752888999989999999999979998805896199689961499814589999999999999999999999999999999834478876303202300112024899999999988999999999973244037999999999999999996989999981998999999985887699 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDNASFSEPWPANASGPDPALSCSNASTLAPLPAPLAVAVPVVYAVICAVGLAGNSAVLYVLLRAPRMKTVTNLFILNLAIADELFTLVLPINIADFLLRQWPFGELMCKLIVAIDQYNTFSSLYFLTVMSADRYLVVLATAESRRVAGRTYSAARAVSLAVWGIVTLVVLPFAVFARLDDEQGRRQCVLVFPQPEAFWWRASRLYTLVLGFAIPVSTICVLYTTLLCRLHAMRLDSHAKALERAKKRVTFLVVAILAVCLLCWTPYHLSTVVALTTDLPQTPLVIAISYFITSLSYANSCLNPFLYAFLDASFRRNLRQLITCRAAA | |
| 6444334442332223333323244324344143000000021013002302321220010002133321000000000020312000000000010026300001000100100032111100000000031001000000203314321210010000101110100010000002036474231010201454310210110111221322112000200000021013134554445444321000000000000000012210000001002304422000001010000003000110102000043014101400313668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCC MDNASFSEPWPANASGPDPALSCSNASTLAPLPAPLAVAVPVVYAVICAVGLAGNSAVLYVLLRAPRMKTVTNLFILNLAIADELFTLVLPINIADFLLRQWPFGELMCKLIVAIDQYNTFSSLYFLTVMSADRYLVVLATAESRRVAGRTYSAARAVSLAVWGIVTLVVLPFAVFARLDDEQGRRQCVLVFPQPEAFWWRASRLYTLVLGFAIPVSTICVLYTTLLCRLHAMRLDSHAKALERAKKRVTFLVVAILAVCLLCWTPYHLSTVVALTTDLPQTPLVIAISYFITSLSYANSCLNPFLYAFLDASFRRNLRQLITCRAAA | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.35 | 0.38 | 0.99 | 3.16 | Download | LANEGKVKEAQAAAEQAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALD--FRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG- | |||||||||||||||||||
| 2 | 4n6hA | 0.37 | 0.36 | 0.92 | 4.21 | Download | ------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDF--RTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG- | |||||||||||||||||||
| 3 | 4n6hA | 0.37 | 0.36 | 0.92 | 4.19 | Download | ------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA--LDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG- | |||||||||||||||||||
| 4 | 4djh | 0.36 | 0.33 | 0.87 | 1.56 | Download | --------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDF--RTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---A--LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---- | |||||||||||||||||||
| 5 | 4djh | 0.36 | 0.33 | 0.87 | 1.17 | Download | --------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALD--FRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA-----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---- | |||||||||||||||||||
| 6 | 4n6hA | 0.37 | 0.36 | 0.91 | 3.73 | Download | --------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDF--RTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG- | |||||||||||||||||||
| 7 | 4djh | 0.36 | 0.33 | 0.86 | 1.72 | Download | ----------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRT--PLKAKIINICIWLLSSSVGISAIVLGGTKEDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF----- | |||||||||||||||||||
| 8 | 4n6hA | 0.37 | 0.36 | 0.92 | 4.90 | Download | -----------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPV--KALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG- | |||||||||||||||||||
| 9 | 4n6hA | 0.37 | 0.36 | 0.92 | 3.50 | Download | ------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKAL--DFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG- | |||||||||||||||||||
| 10 | 5c1mA | 0.35 | 0.32 | 0.90 | 5.19 | Download | ---------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKA--LDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
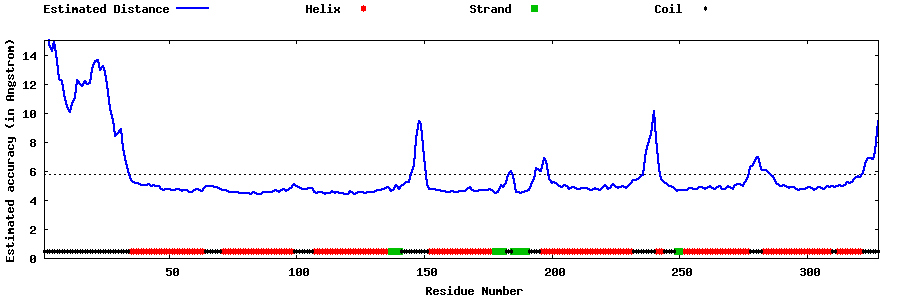
| Generated 3D models | Estimated local accuracy of models | ||
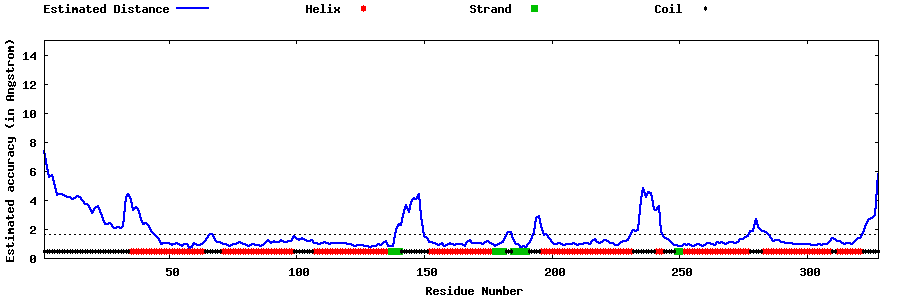 | |||
|
|
|||
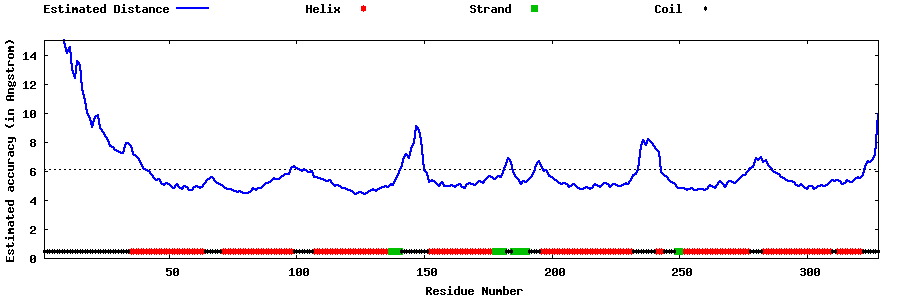 | |||
|
| |||
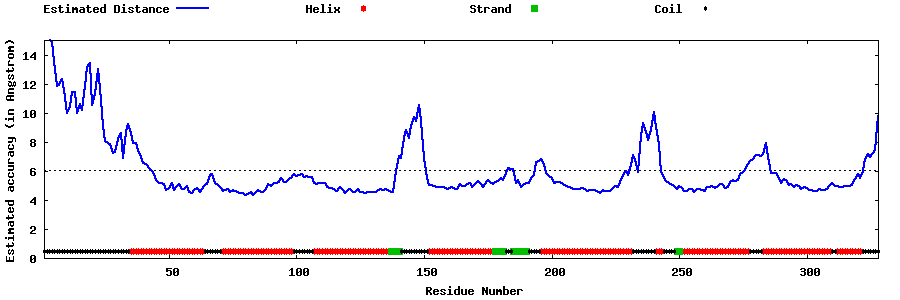 | |||
|
| |||
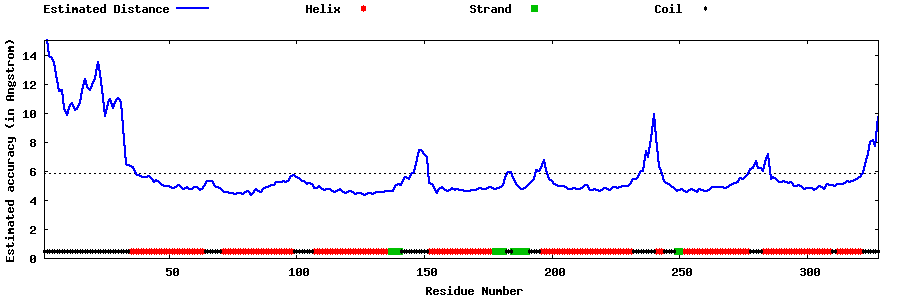 | |||
|
| |||
 | |||
|
| |||