

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MDLPVNLTSFSLSTPSPLETNHSLGKDDLRPSSPLLSVFGVLILTLLGFLVAATFAWNLLVLATILRVRTFHRVPHNLVASMAVSDVLVAALVMPLSLVHELSGRRWQLGRRLCQLWIACDVLCCTASIWNVTAIALDRYWSITRHMEYTLRTRKCVSNVMIALTWALSAVISLAPLLFGWGETYSEGSEECQVSREPSYAVFSTVGAFYLPLCVVLFVYWKIYKAAKFRVGSRKTNSVSPISEAVEVKDSAKQPQMVFTVRHATVTFQPEGDTWREQKEQRAALMVGILIGVFVLCWIPFFLTELISPLCSCDIPAIWKSIFLWLGYSNSFFNPLIYTAFNKNYNSAFKNFFSRQH | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHCHHHHHHCCHHHHHHHHHHHCCCC | |
| 999978887767999877778888877789888750699999999999999999998988378677547888577999999999999999998559999998379244868899999999999999999999999986677434520388746489999978999999999999999967787578999967987361699999999999999999999999999999998775402332211233322221112322212322210000222223323467898999999999999789999999999809988789999999999988613208478378999999999817689 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MDLPVNLTSFSLSTPSPLETNHSLGKDDLRPSSPLLSVFGVLILTLLGFLVAATFAWNLLVLATILRVRTFHRVPHNLVASMAVSDVLVAALVMPLSLVHELSGRRWQLGRRLCQLWIACDVLCCTASIWNVTAIALDRYWSITRHMEYTLRTRKCVSNVMIALTWALSAVISLAPLLFGWGETYSEGSEECQVSREPSYAVFSTVGAFYLPLCVVLFVYWKIYKAAKFRVGSRKTNSVSPISEAVEVKDSAKQPQMVFTVRHATVTFQPEGDTWREQKEQRAALMVGILIGVFVLCWIPFFLTELISPLCSCDIPAIWKSIFLWLGYSNSFFNPLIYTAFNKNYNSAFKNFFSRQH | |
| 873434223232333442433432445424344222101100100211320330332120000000223301100000000001001100000023100000242302103100000000000000000300000010000000200314332232000000000022012102100000023334643420202333100000001003200200020000002003323433443433434444434334433333333434223332322222344343012000000010120021000100020004130032000000120130012000000100420030023104358 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHCHHHHHHCCHHHHHHHHHHHCCCC MDLPVNLTSFSLSTPSPLETNHSLGKDDLRPSSPLLSVFGVLILTLLGFLVAATFAWNLLVLATILRVRTFHRVPHNLVASMAVSDVLVAALVMPLSLVHELSGRRWQLGRRLCQLWIACDVLCCTASIWNVTAIALDRYWSITRHMEYTLRTRKCVSNVMIALTWALSAVISLAPLLFGWGETYSEGSEECQVSREPSYAVFSTVGAFYLPLCVVLFVYWKIYKAAKFRVGSRKTNSVSPISEAVEVKDSAKQPQMVFTVRHATVTFQPEGDTWREQKEQRAALMVGILIGVFVLCWIPFFLTELISPLCSCDIPAIWKSIFLWLGYSNSFFNPLIYTAFNKNYNSAFKNFFSRQH | |||||||||||||||||||||||||
| 1 | 3sn6R | 0.30 | 0.26 | 0.78 | 2.66 | Download | ------------------------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTK-TWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRNCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR------------------------------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC--- | |||||||||||||||||||
| 2 | 4ib4A | 0.25 | 0.28 | 0.89 | 4.55 | Download | ---------------------------------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNY | |||||||||||||||||||
| 3 | 4iaqA | 0.39 | 0.33 | 0.78 | 3.44 | Download | ----------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR-WTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPP------FFWRQASECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSR--------------------------------------IIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI-----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK- | |||||||||||||||||||
| 4 | 4ib4 | 0.27 | 0.28 | 0.90 | 1.56 | Download | --------------------------EEQGNK---L----HWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIET--NPNNITCVLTKEGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNADAAQVAAQKKDFRHGGKVKEAQKNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNY | |||||||||||||||||||
| 5 | 3uon | 0.24 | 0.29 | 0.89 | 1.23 | Download | ------------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIG-YWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVRTVEDGECYIQFNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLGNAAGNTNGVITNAKAGRVITTFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--- | |||||||||||||||||||
| 6 | 3sn6R | 0.30 | 0.26 | 0.78 | 2.95 | Download | -------------------------------------VWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKT-WTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEG------------------------------------RCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC--- | |||||||||||||||||||
| 7 | 4ib4 | 0.26 | 0.28 | 0.88 | 1.78 | Download | ----------------------------------------HWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETN--PNNITCVLTKEGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVMRAAAQKGFGKVKEEQTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCN- | |||||||||||||||||||
| 8 | 2rh1A | 0.28 | 0.29 | 0.89 | 4.17 | Download | --------------------------------DE---VWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMK-MWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLGVAGFTNSLRMLQQKRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTGKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLCL-- | |||||||||||||||||||
| 9 | 4iaqA | 0.39 | 0.33 | 0.78 | 2.86 | Download | ----------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTG-RWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWR-------QASECVVNTHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRI--------------------------------------IQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI-----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK- | |||||||||||||||||||
| 10 | 3zpqA | 0.32 | 0.28 | 0.78 | 3.40 | Download | ------------------------------GAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRG-TWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQ------------------------------------------SRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRLLA--- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
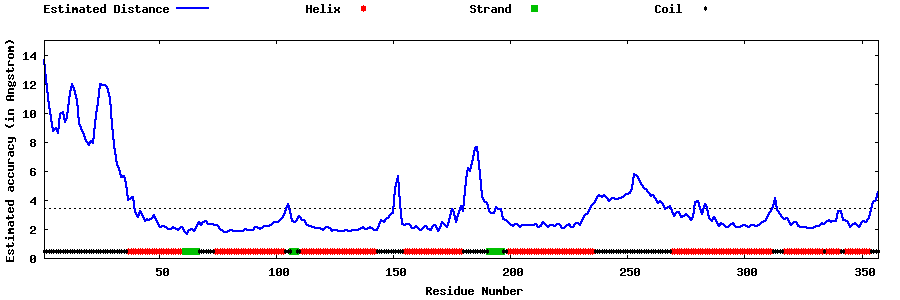 | |||
|
|
|||
 | |||
|
| |||
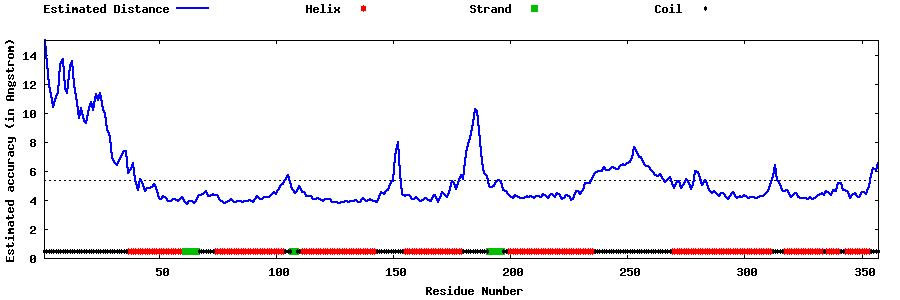 | |||
|
| |||
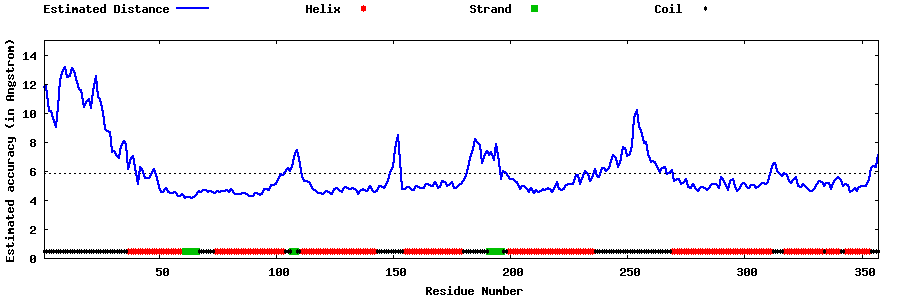 | |||
|
| |||
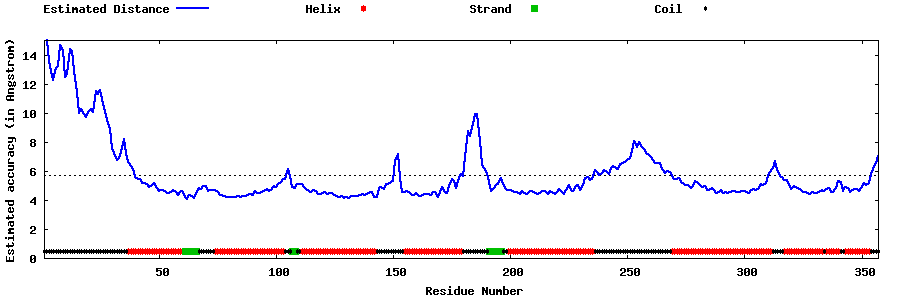 | |||
|
| |||