

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNEDLKVNLSGLPRDYLDAAAAENISAAVSSRVPAVEPEPELVVNPWDIVLCTSGTLISCENAIVVLIIFHNPSLRAPMFLLIGSLALADLLAGIGLITNFVFAYLLQSEATKLVTIGLIVASFSASVCSLLAITVDRYLSLYYALTYHSERTVTFTYVMLVMLWGTSICLGLLPVMGWNCLRDESTCSVVRPLTKNNAAILSVSFLFMFALMLQLYIQICKIVMRHAHQIALQHHFLATSHYVTTRKGVSTLAIILGTFAACWMPFTLYSLIADYTYPSIYTYATLLPATYNSIINPVIYAFRNQEIQKALCLICCGCIPSSLAQRARSPSDV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSSCCCCSSSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCC | |
| 9987788988687000587767888877567899889985449999999999999999999998634226458788699999999999999999999999999985486899999999999999999999999999728884352556667748888888789999999999999998076158870588740556708999986237999999999999999999861214666653204677786999999999999999999999999999968928999999999999998989999928999999999996699899988889999999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNEDLKVNLSGLPRDYLDAAAAENISAAVSSRVPAVEPEPELVVNPWDIVLCTSGTLISCENAIVVLIIFHNPSLRAPMFLLIGSLALADLLAGIGLITNFVFAYLLQSEATKLVTIGLIVASFSASVCSLLAITVDRYLSLYYALTYHSERTVTFTYVMLVMLWGTSICLGLLPVMGWNCLRDESTCSVVRPLTKNNAAILSVSFLFMFALMLQLYIQICKIVMRHAHQIALQHHFLATSHYVTTRKGVSTLAIILGTFAACWMPFTLYSLIADYTYPSIYTYATLLPATYNSIINPVIYAFRNQEIQKALCLICCGCIPSSLAQRARSPSDV | |
| 7475343423212332233433322223234433434331200000111111100220332330000000103300100000000001000010111112001000130310021110000000000130000001000000020020333123300000000001201320100000000244442000000133310010123233322211203120010003003302434644444433311100000000011121131301000000111123000000013013010120010001043003000200012123645444534366 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSSCCCCSSSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCC MNEDLKVNLSGLPRDYLDAAAAENISAAVSSRVPAVEPEPELVVNPWDIVLCTSGTLISCENAIVVLIIFHNPSLRAPMFLLIGSLALADLLAGIGLITNFVFAYLLQSEATKLVTIGLIVASFSASVCSLLAITVDRYLSLYYALTYHSERTVTFTYVMLVMLWGTSICLGLLPVMGWNCLRDESTCSVVRPLTKNNAAILSVSFLFMFALMLQLYIQICKIVMRHAHQIALQHHFLATSHYVTTRKGVSTLAIILGTFAACWMPFTLYSLIADYTYPSIYTYATLLPATYNSIINPVIYAFRNQEIQKALCLICCGCIPSSLAQRARSPSDV | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.26 | 0.25 | 0.84 | 3.20 | Download | ----------------------ENFMD-----IECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHI-DETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRA------MSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 2 | 4z34A | 0.26 | 0.28 | 0.89 | 3.88 | Download | -----------------QCFYNESIAFFYNRSGKHLATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQ-LHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFAIFNLVTFVVMVVLYAHIFGYVADLEDNWENAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQIL------------------ | |||||||||||||||||||
| 3 | 5tgzA | 0.26 | 0.25 | 0.85 | 3.48 | Download | -----------------------GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHI-DKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA---------PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 4 | 4ib4 | 0.17 | 0.20 | 0.85 | 1.54 | Download | -------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFGDFLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 5 | 4ib4 | 0.17 | 0.20 | 0.85 | 1.18 | Download | ------------------------------------EEQ-GNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 6 | 5tjvA | 0.26 | 0.25 | 0.84 | 3.16 | Download | ----------------------ENFMD-----IECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHI-DETYLMFWIGVTSVLLLFIVYAYMYILWKA------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 7 | 4ib4 | 0.17 | 0.20 | 0.83 | 1.73 | Download | ---------------------------------------------WAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFGFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDQQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 8 | 4bvnA | 0.23 | 0.23 | 0.82 | 3.80 | Download | --------------------------------------LSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRK-----RKTSRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNVPKWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLLA----------------- | |||||||||||||||||||
| 9 | 5tjvA | 0.25 | 0.25 | 0.84 | 3.33 | Download | ---------------------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFP-HIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSF------SDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 10 | 5g53A | 0.26 | 0.24 | 0.81 | 3.99 | Download | -------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNCGEGQVACLFVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMT----------LQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLENLY--------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
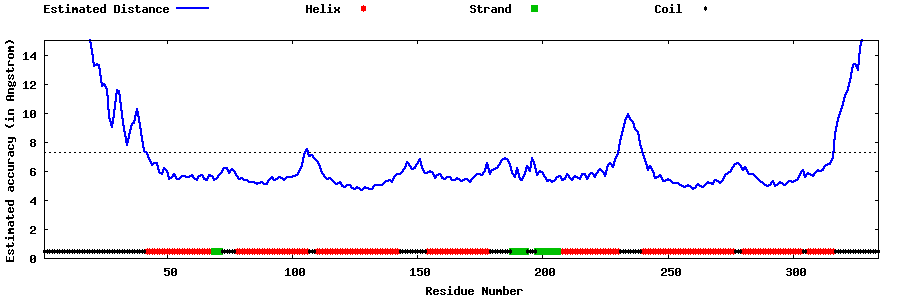
| Generated 3D models | Estimated local accuracy of models | ||
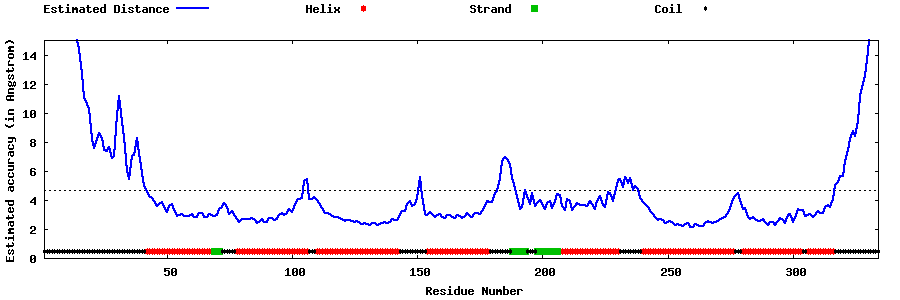 | |||
|
|
|||
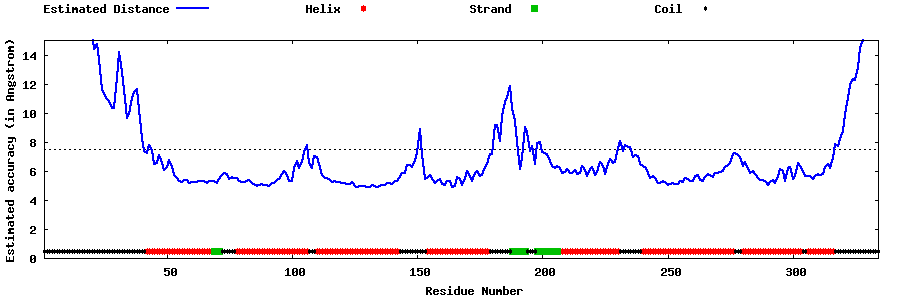 | |||
|
| |||
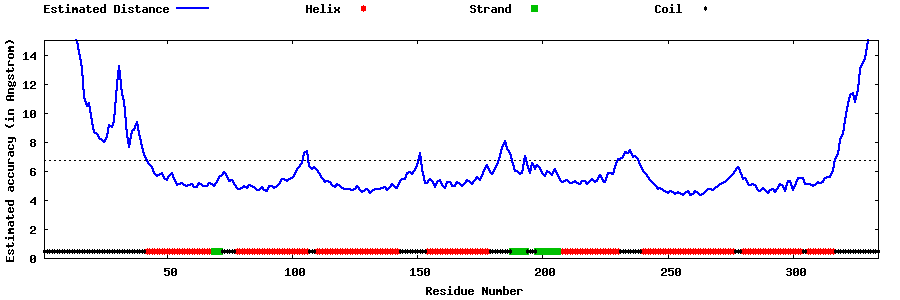 | |||
|
| |||
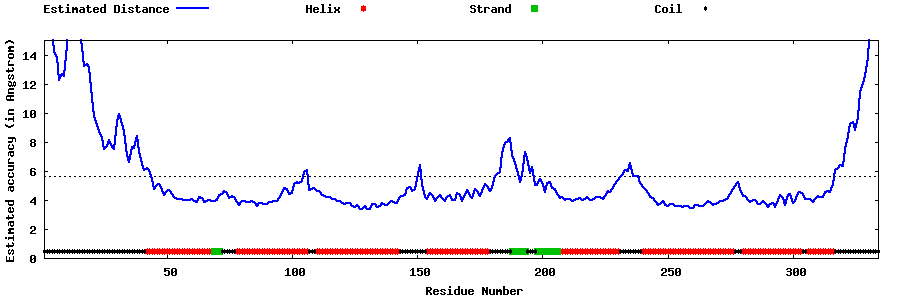 | |||
|
| |||
 | |||
|
| |||