

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MELAVGNLSEGNASWPEPPAPEPGPLFGIGVENFVTLVVFGLIFALGVLGNSLVITVLARSKPGKPRSTTNLFILNLSIADLAYLLFCIPFQATVYALPTWVLGAFICKFIHYFFTVSMLVSIFTLAAMSVDRYVAIVHSRRSSSLRVSRNALLGVGCIWALSIAMASPVAYHQGLFHPRASNQTFCWEQWPDPRHKKAYVVCTFVFGYLLPLLLICFCYAKVLNHLHKKLKNMSKKSEASKKKTAQTVLVVVVVFGISWLPHHIIHLWAEFGVFPLTPASFLFRITAHCLAYSNSSVNPIIYAFLSENFRKAYKQVFKCHIRKDSHLSDTKESKSRIDTPPSTNCTHV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9888889888888888988665666444139999999999999999865588720020121888777699999999999999999995889999998489879538888899999999999999999999982865302686010420564864404999999999989999826888379980687087995579999999999999999999999999999999951278622456763002545788999999999889999999999544457507889999999999999959999999809999999999946002578777888789877778998987789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MELAVGNLSEGNASWPEPPAPEPGPLFGIGVENFVTLVVFGLIFALGVLGNSLVITVLARSKPGKPRSTTNLFILNLSIADLAYLLFCIPFQATVYALPTWVLGAFICKFIHYFFTVSMLVSIFTLAAMSVDRYVAIVHSRRSSSLRVSRNALLGVGCIWALSIAMASPVAYHQGLFHPRASNQTFCWEQWPDPRHKKAYVVCTFVFGYLLPLLLICFCYAKVLNHLHKKLKNMSKKSEASKKKTAQTVLVVVVVFGISWLPHHIIHLWAEFGVFPLTPASFLFRITAHCLAYSNSSVNPIIYAFLSENFRKAYKQVFKCHIRKDSHLSDTKESKSRIDTPPSTNCTHV | |
| 2423231313312323333333424223220000000211120011013112000000000335401100010020003001000200110100100243011020001000001110000000000000200020001002123321221000000102210110010100002023363442201103012630320000010331123101000200000011013334434444444220000000000000000021200000021023232321000010000000010001000000000430141024003022345454544444444344444444435 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MELAVGNLSEGNASWPEPPAPEPGPLFGIGVENFVTLVVFGLIFALGVLGNSLVITVLARSKPGKPRSTTNLFILNLSIADLAYLLFCIPFQATVYALPTWVLGAFICKFIHYFFTVSMLVSIFTLAAMSVDRYVAIVHSRRSSSLRVSRNALLGVGCIWALSIAMASPVAYHQGLFHPRASNQTFCWEQWPDPRHKKAYVVCTFVFGYLLPLLLICFCYAKVLNHLHKKLKNMSKKSEASKKKTAQTVLVVVVVFGISWLPHHIIHLWAEFGVFPLTPASFLFRITAHCLAYSNSSVNPIIYAFLSENFRKAYKQVFKCHIRKDSHLSDTKESKSRIDTPPSTNCTHV | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.29 | 0.30 | 0.91 | 2.85 | Download | AQAAAEQLKTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYT--KMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.31 | 0.30 | 0.82 | 3.88 | Download | ------------------------------ALAIAITALYSAVCAVGLLGNVLVMFGIVRYT--KMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR--DGAVVCMLQFPSPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP---------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.30 | 0.30 | 0.85 | 3.59 | Download | --------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYT--KMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.32 | 0.30 | 0.81 | 1.55 | Download | ----------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYT--KMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA--A---LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.32 | 0.30 | 0.81 | 1.18 | Download | ----------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYT--KMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.30 | 0.30 | 0.85 | 3.43 | Download | ----------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYT--KMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.21 | 0.20 | 0.80 | 1.71 | Download | --------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNR--HLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGRTVEDGECYIQFFSN---AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC---IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------ | |||||||||||||||||||
| 8 | 4ea3A | 0.33 | 0.29 | 0.79 | 4.86 | Download | ----------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHT--KMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPT-------SSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEEIECLVEIPTPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGRLLSGSREKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR---------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.29 | 0.30 | 0.85 | 3.24 | Download | --------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYT--KMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.30 | 0.30 | 0.91 | 3.92 | Download | LKLANEGKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMKTATNIYIFNLALADALATS-TLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
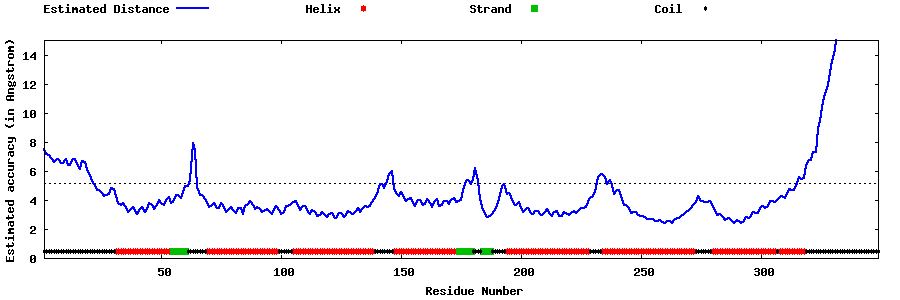 | |||
|
|
|||
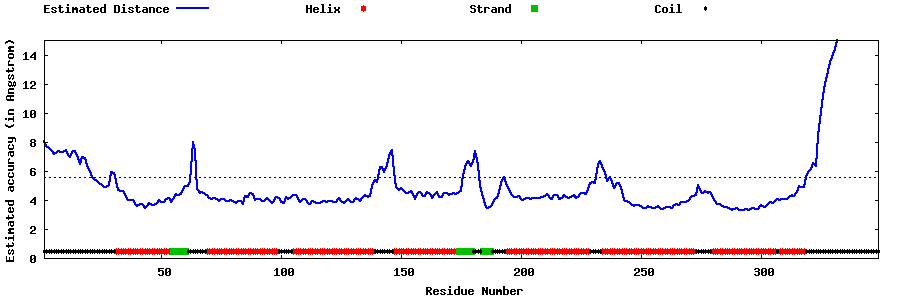 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||