

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNASAASLNDSQVVVVAAEGAAAAATAAGGPDTGEWGPPAAAALGAGGGANGSLELSSQLSAGPPGLLLPAVNPWDVLLCVSGTVIAGENALVVALIASTPALRTPMFVLVGSLATADLLAGCGLILHFVFQYLVPSETVSLLTVGFLVASFAASVSSLLAITVDRYLSLYNALTYYSRRTLLGVHLLLAATWTVSLGLGLLPVLGWNCLAERAACSVVRPLARSHVALLSAAFFMVFGIMLHLYVRICQVVWRHAHQIALQQHCLAPPHLAATRKGVGTLAVVLGTFGASWLPFAIYCVVGSHEDPAVYTYATLLPATYNSMINPIIYAFRNQEIQRALWLLLCGCFQSKVPFRSRSPSEV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCC | |
| 98777897877664442013334477678998766799854556888888888676778889984224308899999999999999998998311013578889789999999999999999999999999996085079999999999999999999999999738855863568876559999999999999999999999725676578883000101663304411269999999999999999999999987750455543332111231024456899999999988898873021247834999999999997887858999728999999999997255789999888899999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNASAASLNDSQVVVVAAEGAAAAATAAGGPDTGEWGPPAAAALGAGGGANGSLELSSQLSAGPPGLLLPAVNPWDVLLCVSGTVIAGENALVVALIASTPALRTPMFVLVGSLATADLLAGCGLILHFVFQYLVPSETVSLLTVGFLVASFAASVSSLLAITVDRYLSLYNALTYYSRRTLLGVHLLLAATWTVSLGLGLLPVLGWNCLAERAACSVVRPLARSHVALLSAAFFMVFGIMLHLYVRICQVVWRHAHQIALQQHCLAPPHLAATRKGVGTLAVVLGTFGASWLPFAIYCVVGSHEDPAVYTYATLLPATYNSMINPIIYAFRNQEIQRALWLLLCGCFQSKVPFRSRSPSEV | |
| 75244342334423323333233233333344334332333232333432232132445244433313122110000000000120131120000000103300000000000000000200211022001000230310030100000000000230000000000000020030433022300000000011101210100000012234432000000233200000012133112100200110010013103303434544444333331100000000000000000000000000122200000000012013311220003002143003001200000034634443333456 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCC MNASAASLNDSQVVVVAAEGAAAAATAAGGPDTGEWGPPAAAALGAGGGANGSLELSSQLSAGPPGLLLPAVNPWDVLLCVSGTVIAGENALVVALIASTPALRTPMFVLVGSLATADLLAGCGLILHFVFQYLVPSETVSLLTVGFLVASFAASVSSLLAITVDRYLSLYNALTYYSRRTLLGVHLLLAATWTVSLGLGLLPVLGWNCLAERAACSVVRPLARSHVALLSAAFFMVFGIMLHLYVRICQVVWRHAHQIALQQHCLAPPHLAATRKGVGTLAVVLGTFGASWLPFAIYCVVGSHEDPAVYTYATLLPATYNSMINPIIYAFRNQEIQRALWLLLCGCFQSKVPFRSRSPSEV | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.25 | 0.24 | 0.78 | 2.69 | Download | --------------------------------------------------ENFMDIECFMV-----LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPH-IDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAM------SFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 2 | 5uenA | 0.24 | 0.26 | 0.77 | 3.79 | Download | -----------------------------------------------------------------------QAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINTYFH--TCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSAKCEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDNTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRCQ----------- | |||||||||||||||||||
| 3 | 5tgzA | 0.27 | 0.25 | 0.78 | 3.13 | Download | ---------------------------------------------------GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHI-DKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA---------PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 4 | 4ib4 | 0.18 | 0.21 | 0.78 | 1.55 | Download | -----------------------------------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFGDFLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 5 | 4ib4 | 0.18 | 0.21 | 0.78 | 1.18 | Download | -----------------------------------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 6 | 5tjvA | 0.25 | 0.24 | 0.78 | 2.91 | Download | --------------------------------------------------------NFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHI-DETYLMFWIGVTSVLLLFIVYAYMYILWKA------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 7 | 4ib4 | 0.19 | 0.21 | 0.75 | 1.75 | Download | ---------------------------------------------------------------------------ALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDQQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNY-------------- | |||||||||||||||||||
| 8 | 4bvnA | 0.25 | 0.24 | 0.75 | 3.07 | Download | ------------------------------------------------------------------LSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRK-----RKTSRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRPKWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLLA----------------- | |||||||||||||||||||
| 9 | 5tjvA | 0.26 | 0.24 | 0.78 | 2.92 | Download | -------------------------------------------------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 10 | 5g53A | 0.24 | 0.23 | 0.75 | 2.95 | Download | -----------------------------------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCCGEGACLFEDVVPMNYMVNFFACVLVPLLLMLGVYLRIFLAARRQLKQM----------TLQKEVHAAKSLAIIVGLFALCWLPLHIINCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLENLY--------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
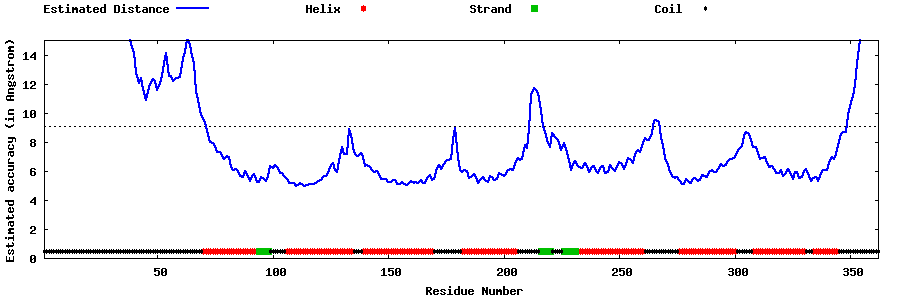
| Generated 3D models | Estimated local accuracy of models | ||
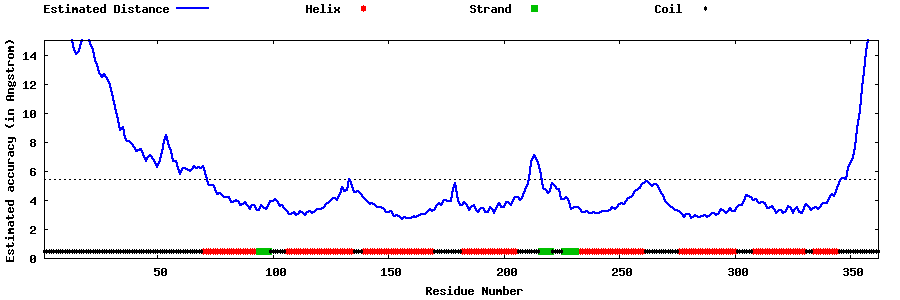 | |||
|
|
|||
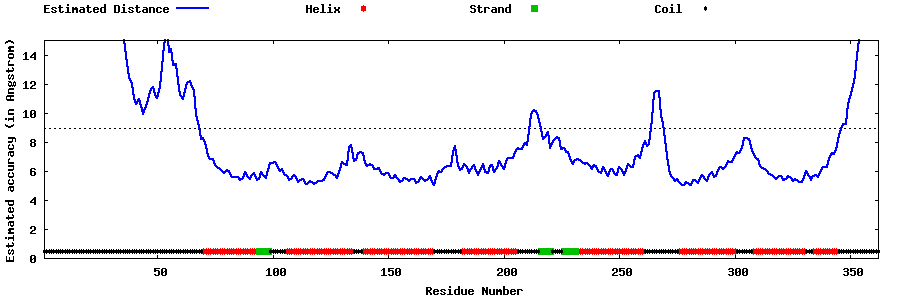 | |||
|
| |||
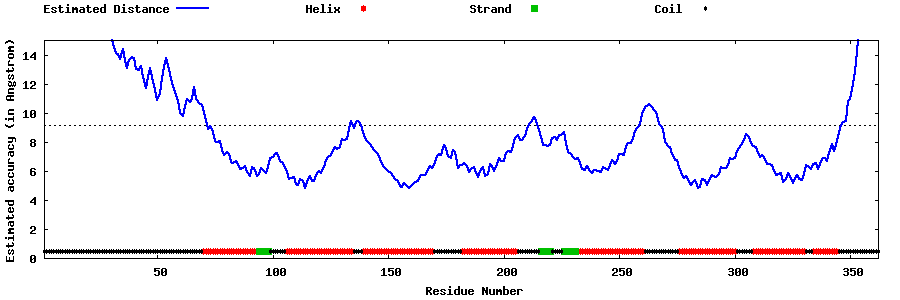 | |||
|
| |||
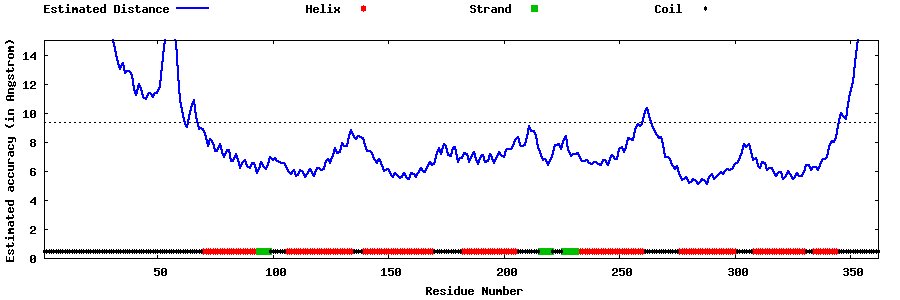 | |||
|
| |||
 | |||
|
| |||