

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MMWGAGSPLAWLSAGSGNVNVSSVGPAEGPTGPAAPLPSPKAWDVVLCISGTLVSCENALVVAIIVGTPAFRAPMFLLVGSLAVADLLAGLGLVLHFAAVFCIGSAEMSLVLVGVLAMAFTASIGSLLAITVDRYLSLYNALTYYSETTVTRTYVMLALVWGGALGLGLLPVLAWNCLDGLTTCGVVYPLSKNHLVVLAIAFFMVFGIMLQLYAQICRIVCRHAQQIALQRHLLPASHYVATRKGIATLAVVLGAFAACWLPFTVYCLLGDAHSPPLYTYLTLLPATYNSMINPIIYAFRNQDVQKVLWAVCCCCSSSKIPFRSRSPSDV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSSCCCCSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCC | |
| 999999996533467899876778887788887777259999999999999999997898300035168888699999999999999999999999999997595039889989999999999999999999708870554668877168888874369999999999999996687468870256765256358998708999999999999999999999887760344555326677888889888899999999987999999997616977999999999999997809999806999999999994765789999888999999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MMWGAGSPLAWLSAGSGNVNVSSVGPAEGPTGPAAPLPSPKAWDVVLCISGTLVSCENALVVAIIVGTPAFRAPMFLLVGSLAVADLLAGLGLVLHFAAVFCIGSAEMSLVLVGVLAMAFTASIGSLLAITVDRYLSLYNALTYYSETTVTRTYVMLALVWGGALGLGLLPVLAWNCLDGLTTCGVVYPLSKNHLVVLAIAFFMVFGIMLQLYAQICRIVCRHAQQIALQRHLLPASHYVATRKGIATLAVVLGAFAACWLPFTVYCLLGDAHSPPLYTYLTLLPATYNSMINPIIYAFRNQDVQKVLWAVCCCCSSSKIPFRSRSPSDV | |
| 713344332331323233222322444533433213223210011011110220132120000000113300100000000001000110222132201000133110000000001000100230000001000000020030333033300000000001200210210000002264342000000233200110023133212100200210010013103303434544644433312100000000000011011300000000102223000000012013311210002002143003001200002235536434444256 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSSCCCCSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCC MMWGAGSPLAWLSAGSGNVNVSSVGPAEGPTGPAAPLPSPKAWDVVLCISGTLVSCENALVVAIIVGTPAFRAPMFLLVGSLAVADLLAGLGLVLHFAAVFCIGSAEMSLVLVGVLAMAFTASIGSLLAITVDRYLSLYNALTYYSETTVTRTYVMLALVWGGALGLGLLPVLAWNCLDGLTTCGVVYPLSKNHLVVLAIAFFMVFGIMLQLYAQICRIVCRHAQQIALQRHLLPASHYVATRKGIATLAVVLGAFAACWLPFTVYCLLGDAHSPPLYTYLTLLPATYNSMINPIIYAFRNQDVQKVLWAVCCCCSSSKIPFRSRSPSDV | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.25 | 0.26 | 0.85 | 2.98 | Download | ----------------ENFMDI-------ECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHDETYLMFWIGVTSVLLLFIVYAYMYILWKAG--------KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 2 | 4z34A | 0.25 | 0.28 | 0.91 | 3.88 | Download | ------------PQCFYNESIAFFYNRSGKHLATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQ-LHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWENAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQIL------------------ | |||||||||||||||||||
| 3 | 5tgzA | 0.25 | 0.27 | 0.86 | 3.36 | Download | ------------GGRGENFMDIECFMVLNP-------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHI-DKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA---------PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 4 | 4ib4 | 0.18 | 0.21 | 0.86 | 1.55 | Download | ---------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFGFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 5 | 4ib4 | 0.18 | 0.21 | 0.86 | 1.18 | Download | ---------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 6 | 5tjvA | 0.25 | 0.26 | 0.85 | 3.07 | Download | --------------------------MDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHI-DETYLMFWIGVTSVLLLFIVYAYMYILWKA------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 7 | 4ib4 | 0.18 | 0.21 | 0.84 | 1.73 | Download | ----------------------------------------HWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFGFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCNQQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 8 | 2z73A | 0.19 | 0.23 | 0.94 | 4.02 | Download | --------------ETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAEGVLCNCSFDYISRDLG-------FFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKEANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFTCCQFDETEDDKDAETEI | |||||||||||||||||||
| 9 | 5tjvA | 0.25 | 0.26 | 0.85 | 3.12 | Download | -----------------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSF--------SDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------ | |||||||||||||||||||
| 10 | 5g53A | 0.25 | 0.25 | 0.82 | 3.73 | Download | ---------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNCGEGQVACLFEDVVPMNYMVYFNFFVLVPLLLMLGVYLRIFLAARRQLKQMT----------LQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLENLY--------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
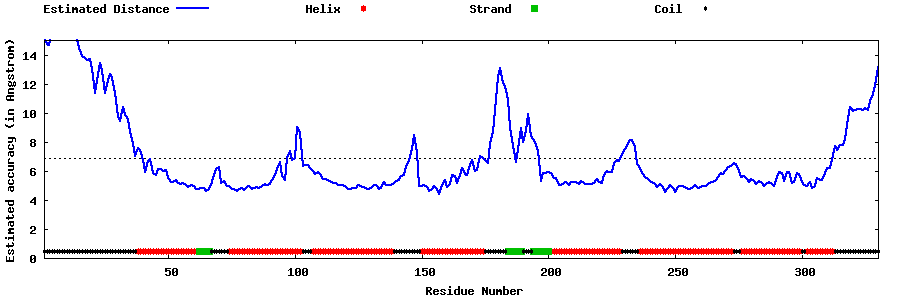
| Generated 3D models | Estimated local accuracy of models | ||
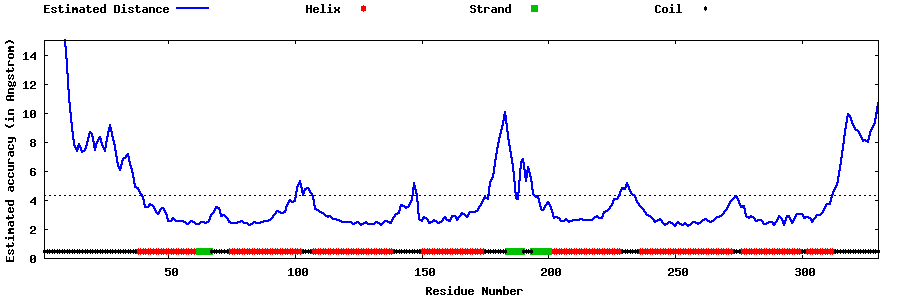 | |||
|
|
|||
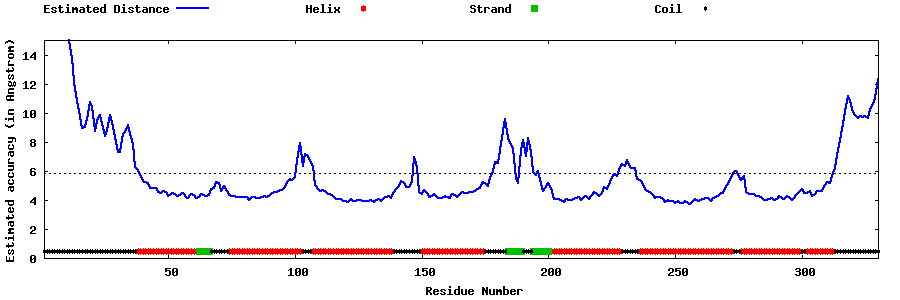 | |||
|
| |||
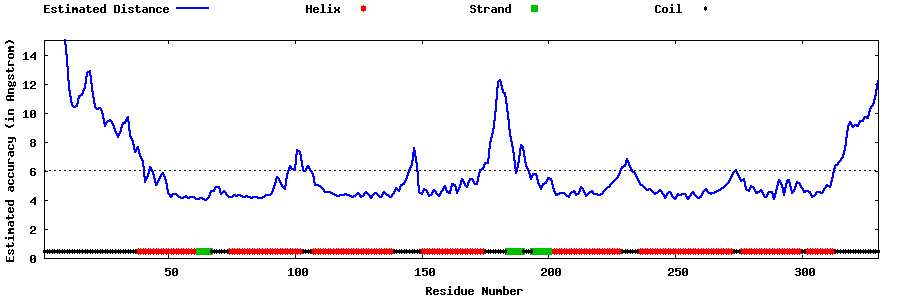 | |||
|
| |||
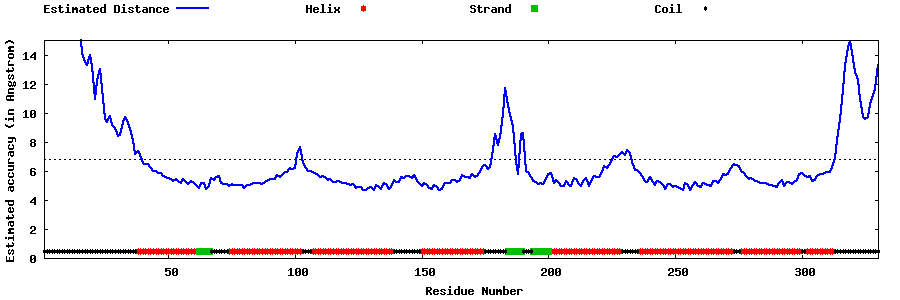 | |||
|
| |||
 | |||
|
| |||