

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MADSCRNLTYVRGSVGPATSTLMFVAGVVGNGLALGILSARRPARPSAFAVLVTGLAATDLLGTSFLSPAVFVAYARNSSLLGLARGGPALCDAFAFAMTFFGLASMLILFAMAVERCLALSHPYLYAQLDGPRCARLALPAIYAFCVLFCALPLLGLGQHQQYCPGSWCFLRMRWAQPGGAAFSLAYAGLVALLVAAIFLCNGSVTLSLCRMYRQQKRHQGSLGPRPRTGEDEVDHLILLALMTVVMAVCSLPLTIRCFTQAVAPDSSSEMGDLLAFRFYAFNPILDPWVFILFRKAVFQRLKLWVCCLCLGPAHGDSQTPLSQLASGRRDPRAPSAPVGKEGSCVPLSAWGEGQVEPLPPTQQSSGSAVGTSSKAEASVACSLC | |
| CCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCSSSSSCCCCCHHHHHHHHSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99888874113778999999999999999999984876610788998318999999999999999988999999970444005436986811029999999999999999999998684601444457772687898999899999999999887565358987799469841689961414363003454799999999999999999999987665113343420456789998987116899999999899999999999788866179999999999999999999999978999999999804378999978877888888788888888888766788877888788777079997678999806888666557778789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MADSCRNLTYVRGSVGPATSTLMFVAGVVGNGLALGILSARRPARPSAFAVLVTGLAATDLLGTSFLSPAVFVAYARNSSLLGLARGGPALCDAFAFAMTFFGLASMLILFAMAVERCLALSHPYLYAQLDGPRCARLALPAIYAFCVLFCALPLLGLGQHQQYCPGSWCFLRMRWAQPGGAAFSLAYAGLVALLVAAIFLCNGSVTLSLCRMYRQQKRHQGSLGPRPRTGEDEVDHLILLALMTVVMAVCSLPLTIRCFTQAVAPDSSSEMGDLLAFRFYAFNPILDPWVFILFRKAVFQRLKLWVCCLCLGPAHGDSQTPLSQLASGRRDPRAPSAPVGKEGSCVPLSAWGEGQVEPLPPTQQSSGSAVGTSSKAEASVACSLC | |
| 73441532322320001111232133133112100000002243342100000000010001001111000000002332012202101000011010001002000100000000000000001203330222100000000012001002102100010123343010000032532110000001133233212000200000000001112324434444444443243002000000000000100000000000000011231220000001022213333100010001330041023002000324444444344444434454453444444463433424333544244244445243322323346434341432 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCSSSSSCCCCCHHHHHHHHSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MADSCRNLTYVRGSVGPATSTLMFVAGVVGNGLALGILSARRPARPSAFAVLVTGLAATDLLGTSFLSPAVFVAYARNSSLLGLARGGPALCDAFAFAMTFFGLASMLILFAMAVERCLALSHPYLYAQLDGPRCARLALPAIYAFCVLFCALPLLGLGQHQQYCPGSWCFLRMRWAQPGGAAFSLAYAGLVALLVAAIFLCNGSVTLSLCRMYRQQKRHQGSLGPRPRTGEDEVDHLILLALMTVVMAVCSLPLTIRCFTQAVAPDSSSEMGDLLAFRFYAFNPILDPWVFILFRKAVFQRLKLWVCCLCLGPAHGDSQTPLSQLASGRRDPRAPSAPVGKEGSCVPLSAWGEGQVEPLPPTQQSSGSAVGTSSKAEASVACSLC | |||||||||||||||||||||||||
| 1 | 4zwjA | 0.16 | 0.18 | 0.96 | 2.47 | Download | VVRSPFEAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLR-TPLNYILLNLAVADLFMVLGGFTSTLYTSLH-----GYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRF-GENHAIMGVAFTWVMALACAAPPLAGWSRYIPEGLQCSCGIDYYTLKPNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQ-------QESATTQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQSCFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKNVIFKKVSRDKSVTIYLGKRDYVDHVSQVEPVDGLVDPELVKGKKVYVTLTCAFRYGQEDIDVMGLTFRRDLYFS | |||||||||||||||||||
| 2 | 2ziy | 0.14 | 0.17 | 0.89 | 1.52 | Download | YNPSIIQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQ-TPANMFIINLAFSDFTFSLVNFPLMTISCFLKKW-----IFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETE---DDK---D--AETEIPAGESSDAAPSA-DAAQMKE--------------------------- | |||||||||||||||||||
| 3 | 1gzmA | 0.17 | 0.18 | 0.76 | 3.61 | Download | -------AEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLR-TPLNYILLNLAVADLFMVFGGFTTTLYTSLH-----GYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFR-FGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETSFVIYMFVVHFIIPLIVIFFCYGQLVFTVKE-------AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKN---DD--------------------------------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.13 | 0.17 | 0.89 | 1.15 | Download | YNPSFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQ-TPANMFIINLAFSDFTFSLVGFPLMTISCF-----LKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAEIPAGESSDAAPSADAAQMKE-------------------------------------- | |||||||||||||||||||
| 5 | 4n6hA | 0.16 | 0.19 | 0.77 | 2.85 | Download | SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK-TATNIYIFNLALADALATST-LPFQSAKYLMET-----WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMA-VTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFVVPILIITVCYGLMLLRLRSV--------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 2ziyA | 0.13 | 0.17 | 0.89 | 2.55 | Download | IVHEFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQ-TPANMFIINLAFSDFTFSLVNFPLMTISCF-----LKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAET----EIPAGESSDAAPSADAAQMKE-------------------------------- | |||||||||||||||||||
| 7 | 4n6hA | 0.15 | 0.19 | 0.76 | 2.78 | Download | --GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK-TATNIYIFNLALADALA-TSTLPFQSAKYLMET-----WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMV-MAVTRPRDGAVVCMLQFPSPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV--------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 2ziyA | 0.12 | 0.17 | 0.89 | 3.40 | Download | HWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQ-TPANMFIINLAFSDFTFSLVNGPLMTISCFL-----KKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETE----DDKDAETEIPAGESSDAAPSADAAQMKE-------------------------------- | |||||||||||||||||||
| 9 | 3uon | 0.11 | 0.20 | 0.74 | 1.72 | Download | ------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQ-TVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYW-----PLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGTVEDGECYIQFFSN---AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEGGAAGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------------------------------------------------------ | |||||||||||||||||||
| 10 | 1u19A | 0.17 | 0.19 | 0.83 | 4.58 | Download | VVRSPFEAPPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLR-TPLNYILLNLAVADLFMVFGGFTTTLYTSLHGY-----FVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRF-GENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETSFVIYMFVVHFIIPLIVIFFCYGQLVFTV-------KEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQ---VAPA------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
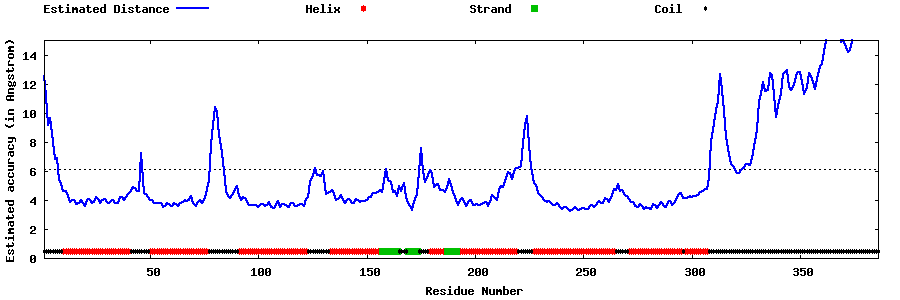
| Generated 3D models | Estimated local accuracy of models | ||
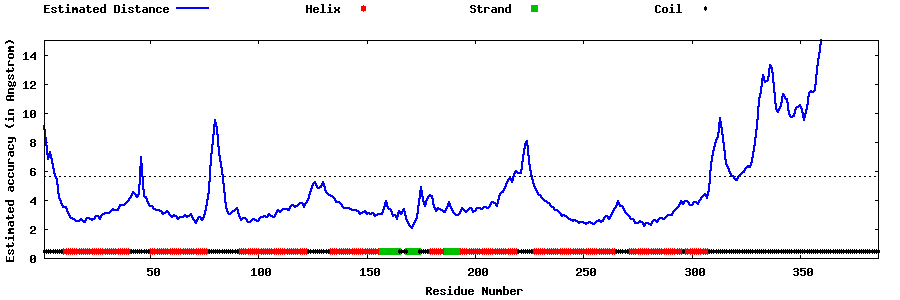 | |||
|
|
|||
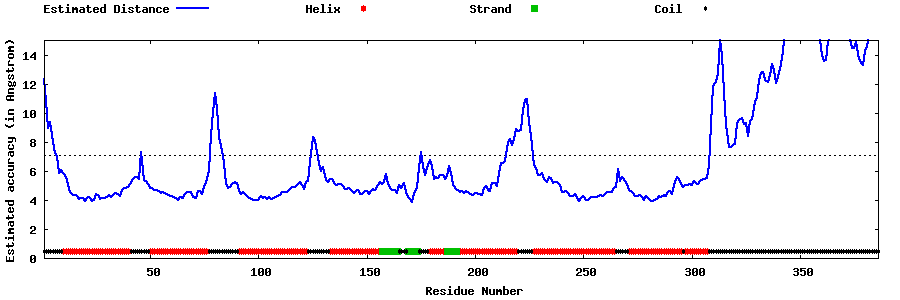 | |||
|
| |||
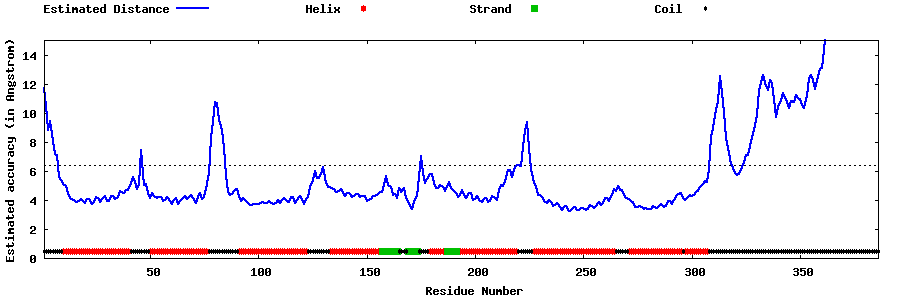 | |||
|
| |||
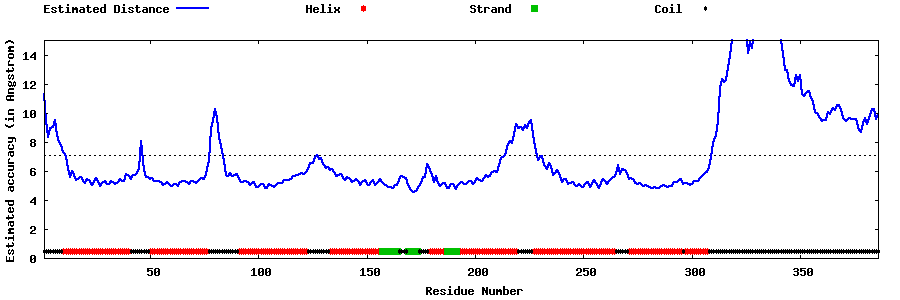 | |||
|
| |||
 | |||
|
| |||