

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNASCCLPSVQPTLPNGSEHLQAPFFSNQSSSAFCEQVFIKPEVFLSLGIVSLLENILVILAVVRNGNLHSPMYFFLCSLAVADMLVSVSNALETIMIAIVHSDYLTFEDQFIQHMDNIFDSMICISLVASICNLLAIAVDRYVTIFYALRYHSIMTVRKALTLIVAIWVCCGVCGVVFIVYSESKMVIVCLITMFFAMMLLMGTLYVHMFLFARLHVKRIAALPPADGVAPQQHSCMKGAVTITILLGVFIFCWAPFFLHLVLIITCPTNPYCICYTAHFNTYLVLIMCNSVIDPLIYAFRSLELRNTFREILCGCNGMNLG | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSCCCHHSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHSSSSSSSSSSSSHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHSSSSSCHHHCHHHHHCCCHHHHHHHHHHHCCCCCCCCC | |
| 99876788889989999988677544468998873068999999999999999988875775777898897899999999999999999999999999986384574566199999999999999999999999999999748850565578877078999999999999999999888833773345621078999999999999999999999999899760477887365642275885658889997200056798867676269997645568787771330240543689998338999999999997389999999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNASCCLPSVQPTLPNGSEHLQAPFFSNQSSSAFCEQVFIKPEVFLSLGIVSLLENILVILAVVRNGNLHSPMYFFLCSLAVADMLVSVSNALETIMIAIVHSDYLTFEDQFIQHMDNIFDSMICISLVASICNLLAIAVDRYVTIFYALRYHSIMTVRKALTLIVAIWVCCGVCGVVFIVYSESKMVIVCLITMFFAMMLLMGTLYVHMFLFARLHVKRIAALPPADGVAPQQHSCMKGAVTITILLGVFIFCWAPFFLHLVLIITCPTNPYCICYTAHFNTYLVLIMCNSVIDPLIYAFRSLELRNTFREILCGCNGMNLG | |
| 44232223323232323333343432424334422331100010111103303322200000002033010000000000010001101311330001003312212130200210020000000000100130000002200000010030333122300000000001100210100002333310000013323311210020021001001300330342455644444333111000000001101000221110000000003333212013000100002013311230103001063003001200000233548 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSCCCHHSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHSSSSSSSSSSSSHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHSSSSSCHHHCHHHHHCCCHHHHHHHHHHHCCCCCCCCC MNASCCLPSVQPTLPNGSEHLQAPFFSNQSSSAFCEQVFIKPEVFLSLGIVSLLENILVILAVVRNGNLHSPMYFFLCSLAVADMLVSVSNALETIMIAIVHSDYLTFEDQFIQHMDNIFDSMICISLVASICNLLAIAVDRYVTIFYALRYHSIMTVRKALTLIVAIWVCCGVCGVVFIVYSESKMVIVCLITMFFAMMLLMGTLYVHMFLFARLHVKRIAALPPADGVAPQQHSCMKGAVTITILLGVFIFCWAPFFLHLVLIITCPTNPYCICYTAHFNTYLVLIMCNSVIDPLIYAFRSLELRNTFREILCGCNGMNLG | |||||||||||||||||||||||||
| 1 | 5tgzA | 0.26 | 0.26 | 0.87 | 2.60 | Download | -------------GGRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-------DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCELQSVCSKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMD-----IELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKL---IKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 2 | 2rh1A | 0.26 | 0.23 | 0.78 | 3.70 | Download | ------------------------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKM------WTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQ----------------LLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNL----IRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLC-------- | |||||||||||||||||||
| 3 | 5tgzA | 0.27 | 0.26 | 0.86 | 3.31 | Download | ----------GGRGENFMDIEC-FMVLNP------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-------VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA----------PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK---LIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.27 | 0.86 | 1.59 | Download | -------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFE--------AMWPLPLVLCPAWLFLDVLASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVIKIETEDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWKIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQ-TTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---- | |||||||||||||||||||
| 5 | 3d4s | 0.23 | 0.26 | 0.83 | 1.16 | Download | ------------------------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMK------MWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIMRATHQEAINCYAQAYASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIWDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNL----IRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL------- | |||||||||||||||||||
| 6 | 5tgzA | 0.26 | 0.26 | 0.87 | 3.06 | Download | --------------GRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-------DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAP-----DQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN---KLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 7 | 4ib4 | 0.19 | 0.27 | 0.85 | 1.75 | Download | -------------------------------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFE-----AMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGETNNNLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWEIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN-QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNY----- | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.18 | 0.84 | 2.38 | Download | ---------------NSD----------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAF----GDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAI-----PTHPGGLVCTPIIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVI---AFVVCWLPYHVRRLMFCYISDEQWTFYMLTNALVYV-----SAAINPILYNLVSANFRQVFLSTL--------- | |||||||||||||||||||
| 9 | 5tgzA | 0.26 | 0.26 | 0.87 | 3.34 | Download | -------------GGRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-------DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCELQSVCSKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARM-----DIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK---LIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 10 | 3emlA | 0.24 | 0.33 | 0.84 | 2.79 | Download | ----------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITI--STGFCAACHGCLFIAC------FVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMEDVVPMNYMVYFNFFACV-----LVPLLLMLGVYLRIFLAARRQLNIFEMLRIDEGLRLKIYKDTEGYYTFALCWLPLHIINCFTFFCPDC--SHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| ||||||||||||||||||||||||||
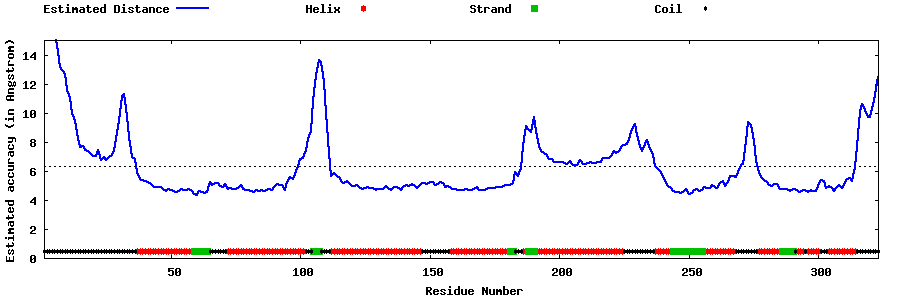
| Generated 3D models | Estimated local accuracy of models | ||
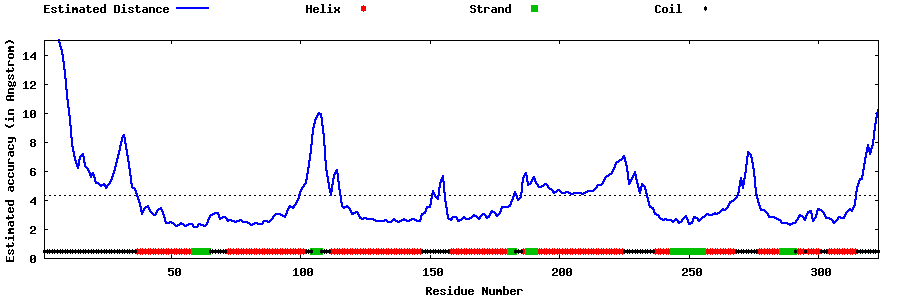 | |||
|
|
|||
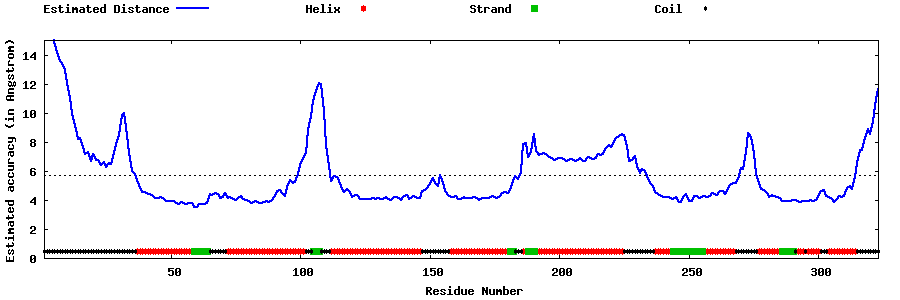 | |||
|
| |||
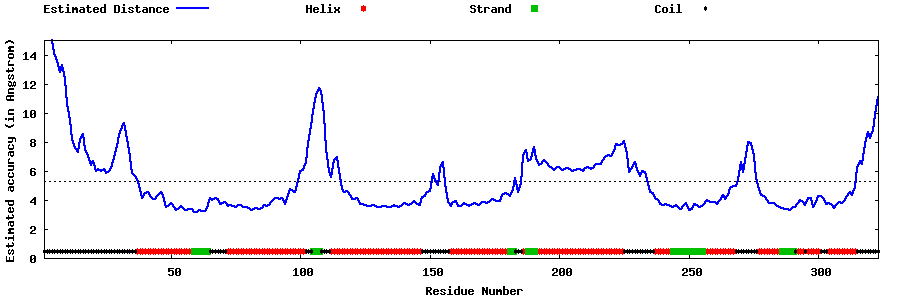 | |||
|
| |||
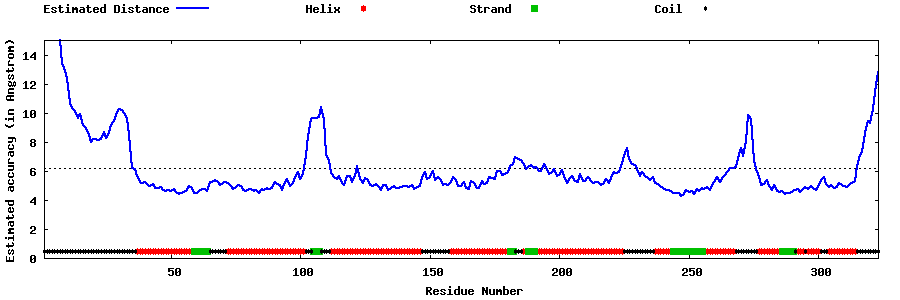 | |||
|
| |||
 | |||
|
| |||