

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MRTLLPPALLTCWLLAPVNSIHPECRFHLEIQEEETKCAELLRSQTEKHKACSGVWDNITCWRPANVGETVTVPCPKVFSNFYSKAGNISKNCTSDGWSETFPDFVDACGYSDPEDESKITFYILVKAIYTLGYSVSLMSLATGSIILCLFRKLHCTRNYIHLNLFLSFILRAISVLVKDDVLYSSSGTLHCPDQPSSWVGCKLSLVFLQYCIMANFFWLLVEGLYLHTLLVAMLPPRRCFLAYLLIGWGLPTVCIGAWTAARLYLEDTGCWDTNDHSVPWWVIRIPILISIIVNFVLFISIIRILLQKLTSPDVGGNDQSQYKRLAKSTLLLIPLFGVHYMVFAVFPISISSKYQILFELCLGSFQGLVVAVLYCFLNSEVQCELKRKWRSRCPTPSASRDYRVCGSSFSRNGSEGALQFHRGSRAQSFLQTETSVI | |
| CCCCCHHHHHHHHCHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCCHHCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCHHHSSSSSSSSCCCCSSSSSSSCCCSSSCCCCCCCCCCCSSSSSSCCCSSSHHHHHHHHHHHSSSSSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 953018999987232031277952599999999999999998717999999987568777589999997798279713057767798333068668816787885444210012333540010023202023208889999999999998722376569999999999999999999999998713221234335567223278999999999999999999998662078772356522128999997243542666650022010123443578853798714631000242244431000023311146664432267789999999999999999999999987621069999999999986887876167761799999999998564789865788882899998999888886879987878677867869 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MRTLLPPALLTCWLLAPVNSIHPECRFHLEIQEEETKCAELLRSQTEKHKACSGVWDNITCWRPANVGETVTVPCPKVFSNFYSKAGNISKNCTSDGWSETFPDFVDACGYSDPEDESKITFYILVKAIYTLGYSVSLMSLATGSIILCLFRKLHCTRNYIHLNLFLSFILRAISVLVKDDVLYSSSGTLHCPDQPSSWVGCKLSLVFLQYCIMANFFWLLVEGLYLHTLLVAMLPPRRCFLAYLLIGWGLPTVCIGAWTAARLYLEDTGCWDTNDHSVPWWVIRIPILISIIVNFVLFISIIRILLQKLTSPDVGGNDQSQYKRLAKSTLLLIPLFGVHYMVFAVFPISISSKYQILFELCLGSFQGLVVAVLYCFLNSEVQCELKRKWRSRCPTPSASRDYRVCGSSFSRNGSEGALQFHRGSRAQSFLQTETSVI | |
| 623000000000010220411333140133035124303520473257331010200010001503113302100021010023431211120233133432110020131334434343222212200000000001000210000010003031321100000000000100010000000112221321232330310000000000001000000000002100100020024430000000000100000000000020112331000125410000002111000022132012100000012034444635324200100100000001022011000000123010010000000003100000100000144005102310342324444446343333333333343333334434442333342236 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCHHHHHHHHCHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCCHHCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCHHHSSSSSSSSCCCCSSSSSSSCCCSSSCCCCCCCCCCCSSSSSSCCCSSSHHHHHHHHHHHSSSSSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MRTLLPPALLTCWLLAPVNSIHPECRFHLEIQEEETKCAELLRSQTEKHKACSGVWDNITCWRPANVGETVTVPCPKVFSNFYSKAGNISKNCTSDGWSETFPDFVDACGYSDPEDESKITFYILVKAIYTLGYSVSLMSLATGSIILCLFRKLHCTRNYIHLNLFLSFILRAISVLVKDDVLYSSSGTLHCPDQPSSWVGCKLSLVFLQYCIMANFFWLLVEGLYLHTLLVAMLPPRRCFLAYLLIGWGLPTVCIGAWTAARLYLEDTGCWDTNDHSVPWWVIRIPILISIIVNFVLFISIIRILLQKLTSPDVGGNDQSQYKRLAKSTLLLIPLFGVHYMVFAVFPISISSKYQILFELCLGSFQGLVVAVLYCFLNSEVQCELKRKWRSRCPTPSASRDYRVCGSSFSRNGSEGALQFHRGSRAQSFLQTETSVI | |||||||||||||||||||||||||
| 1 | 5vaiR | 0.37 | 0.37 | 0.85 | 2.05 | Download | --------------------TVSLSETVQKWREYRRQCQHFLTEALATGLFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWANVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMQHQWDGLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSERIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLM--CKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| 2 | 5vaiA | 0.38 | 0.37 | 0.84 | 4.18 | Download | ----------------------SLSETVQKWREYRRQCQHFLTEAPPTGLFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWANVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWQQHQWDGLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTD--IKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| 3 | 5vaiR | 0.38 | 0.37 | 0.85 | 3.05 | Download | --------------------TVSLSETVQKWREYRRQCQHFLTEAPLATGFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWANVLQGHVYRFCTAEGHWLPKDNSSLPWRDLECEESSPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTAAQQHQYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSERIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIK--CRLAKSTLTLIPLLGTHEVIFAFVMDEHARFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| 4 | 5vai | 0.39 | 0.37 | 0.83 | 2.61 | Download | --------------------TVSLSETVQKWREYRRQCQHFLTEALATGLFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWANVLQGHVYRFCTAEGHWLPKDNDLSECEESSPE-----ERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTLLSYQDS---LGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCK--TDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| 5 | 5vai | 0.38 | 0.37 | 0.85 | 3.20 | Download | --------------------TVSLSETVQKWREYRRQCQHFLTEAPPTGLFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWASNLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECESSPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTAAGLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSERIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTD--IKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| 6 | 5vaiR | 0.39 | 0.37 | 0.85 | 3.26 | Download | ---------------------VSLSETVQKWREYRRQCQHFLTEALATGLFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWASNLQGHVYRFCTAEGHWLPKDNSSLPRDLSECEESSPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTAAQQHQWDDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDI--KCRLAKSTLTLIPLLGTHEVIFAFVMDEHARFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| 7 | 5vai | 0.38 | 0.37 | 0.84 | 4.55 | Download | -----------------------LSETVQKWREYRRQCQHFLTEAPPTGLFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWASNLQGHVYRFCTAEGHWLPKDNSLPWRDLSECEESSPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTAAQLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTD--IKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| 8 | 4k5yA | 0.34 | 0.20 | 0.56 | 3.12 | Download | -------------------------------------------------------------------------------------------------------------------------HYHVAAIINYLGHCISLVALLVAFVLFLRARSIRCLRNIIHANLIAAFILRNATWFVVQLTMS-------PEVHQSNVGWCRLVTAAYNYFHVTNFFWMFGEGCYLHTAIVL--TDRLRAWMFICIGWGVPFPIIVAWAIGKLYYDNEKCWAGKRPGVYDYIYQGPMALVLLINFIFLFNIVRILMTK--LRASTTSETIQARKAVKATLVLLPLLGITYMLAFV--NEVSRVVFIYFNAFLESFQGFFVSVFACFLNS---------------------------------------------------------- | |||||||||||||||||||
| 9 | 5vaiR | 0.38 | 0.37 | 0.85 | 2.68 | Download | --------------------TVSLSETVQKWREYRRQCQHFLTEAPPTGLFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWASVLQGHVYRFCTAEGHWLPKDNSSLWRDLSECEESSPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMQHQWDGLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLM--CKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| 10 | 5vaiR | 0.38 | 0.37 | 0.85 | 1.83 | Download | --------------------TVSLSETVQKWREYRRQCQHFLTEAPPTGLFCNRTFDDYACWPDGAPGSFVNVSCPWYLPWASNVQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESSPEERLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTAAQQHQWQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTD--IKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR-------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
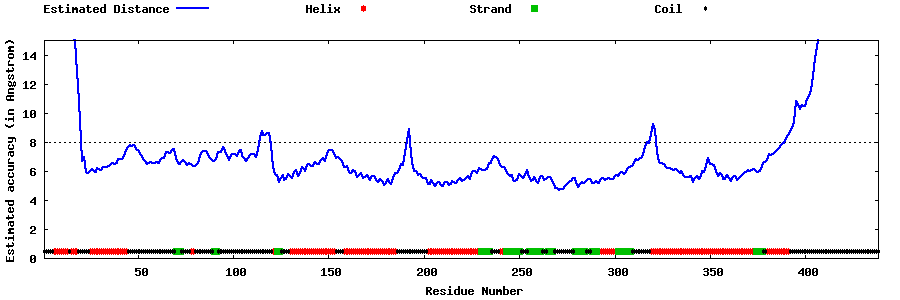
| Generated 3D models | Estimated local accuracy of models | ||
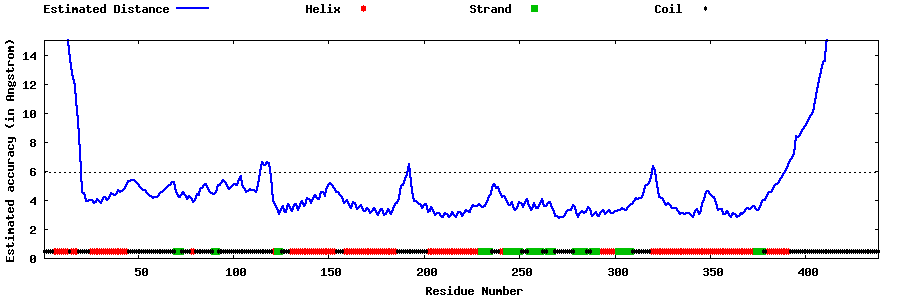 | |||
|
|
|||
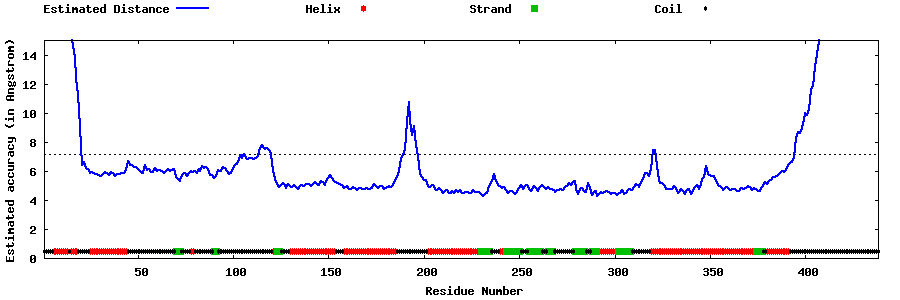 | |||
|
| |||
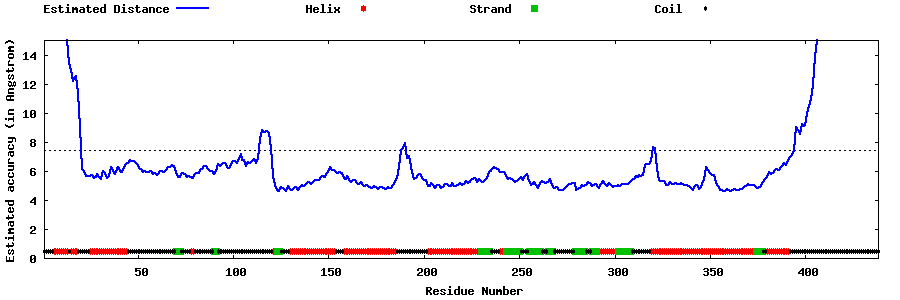 | |||
|
| |||
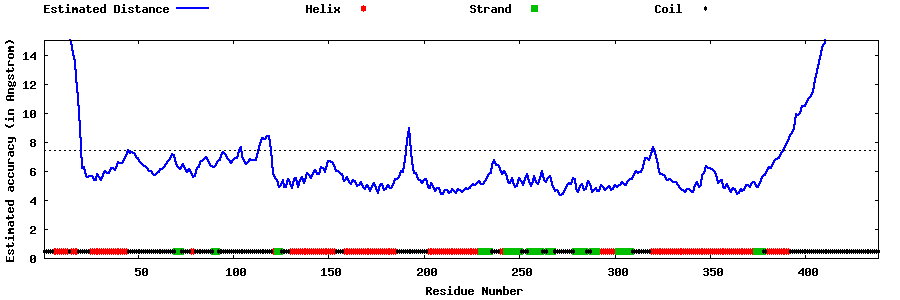 | |||
|
| |||
 | |||
|
| |||