

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MRLSAGPDAGPSGNSSPWWPLATGAGNTSREAEALGEGNGPPRDVRNEELAKLEIAVLAVTFAVAVLGNSSVLLALHRTPRKTSRMHLFIRHLSLADLAVAFFQVLPQMCWDITYRFRGPDWLCRVVKHLQVFGMFASAYMLVVMTADRYIAVCHPLKTLQQPARRSRLMIAAAWVLSFVLSTPQYFVFSMIEVNNVTKARDCWATFIQPWGSRAYVTWMTGGIFVAPVVILGTCYGFICYNIWCNVRGKTASRQSKGAEQAGVAFQKGFLLAPCVSSVKSISRAKIRTVKMTFVIVTAYIVCWAPFFIIQMWSVWDPMSVWTESENPTITITALLGSLNSCCNPWIYMFFSGHLLQDCVQSFPCCQNMKEKFNKEDTDSMSRRQTFYSNNRSPTNSTGMWKDSPKSSKSIKFIPVST | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9899999999998878666666888877788766677888876667500999999999999999999899874753147988852076999999999999999879999999838361504889889999999999999999999987164110678656522699999999999999999999998867787469884578716189813668999999999999999999999999999998733776402321000122211134555544310255788887789999999999999997899999999996665345430799999999999999989999999549999999999845557777677878888743677888889788887888789988778766799999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MRLSAGPDAGPSGNSSPWWPLATGAGNTSREAEALGEGNGPPRDVRNEELAKLEIAVLAVTFAVAVLGNSSVLLALHRTPRKTSRMHLFIRHLSLADLAVAFFQVLPQMCWDITYRFRGPDWLCRVVKHLQVFGMFASAYMLVVMTADRYIAVCHPLKTLQQPARRSRLMIAAAWVLSFVLSTPQYFVFSMIEVNNVTKARDCWATFIQPWGSRAYVTWMTGGIFVAPVVILGTCYGFICYNIWCNVRGKTASRQSKGAEQAGVAFQKGFLLAPCVSSVKSISRAKIRTVKMTFVIVTAYIVCWAPFFIIQMWSVWDPMSVWTESENPTITITALLGSLNSCCNPWIYMFFSGHLLQDCVQSFPCCQNMKEKFNKEDTDSMSRRQTFYSNNRSPTNSTGMWKDSPKSSKSIKFIPVST | |
| 3633432422233221211313333112033333233343334323232001000201020021013001000000002342200000002000200000000001010000003102002000100200100000000000000000001000000202310330000000000010000000000000013266342111000101262020000001012113310110020001000101232433434444443443433344434433334444433312100000000000000000211000000100132234330110000000000010000000000000430040024002003334463445434333334334444443343434354444434434434468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MRLSAGPDAGPSGNSSPWWPLATGAGNTSREAEALGEGNGPPRDVRNEELAKLEIAVLAVTFAVAVLGNSSVLLALHRTPRKTSRMHLFIRHLSLADLAVAFFQVLPQMCWDITYRFRGPDWLCRVVKHLQVFGMFASAYMLVVMTADRYIAVCHPLKTLQQPARRSRLMIAAAWVLSFVLSTPQYFVFSMIEVNNVTKARDCWATFIQPWGSRAYVTWMTGGIFVAPVVILGTCYGFICYNIWCNVRGKTASRQSKGAEQAGVAFQKGFLLAPCVSSVKSISRAKIRTVKMTFVIVTAYIVCWAPFFIIQMWSVWDPMSVWTESENPTITITALLGSLNSCCNPWIYMFFSGHLLQDCVQSFPCCQNMKEKFNKEDTDSMSRRQTFYSNNRSPTNSTGMWKDSPKSSKSIKFIPVST | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.25 | 0.24 | 0.71 | 2.48 | Download | --------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV---------------------------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.17 | 0.20 | 0.79 | 3.85 | Download | -----------------------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDKGSYLRICLLHPVQKFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNIFEMLRIDEGGGSGGDEAEKLFNRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC----------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.25 | 0.24 | 0.72 | 3.47 | Download | --------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL---------------------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.17 | 0.19 | 0.87 | 1.53 | Download | -------DLR---DNETWWYNPSI----IVH-PHWRE-----FDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLE--GVLCNCSFDYISRSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNA--------------KELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEW---VTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVCQFDDKETEDDKDAET-------EIPAGESSDAAPSADAAQMKE-------- | |||||||||||||||||||
| 5 | 4rnb | 0.25 | 0.24 | 0.75 | 1.16 | Download | ----------------------------------------------PKEYEWVLIAGYIIVFVVALIGNVLVCVAVWKNHHMRTVTNYFIVNLSLADVLVTITCLPATLVVDITETWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPST-----AKRARNSIVIIWIVSCIIMIPQAIVMECSTVFKTTLFTVCDERWGGEIYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSFWNESKLSDEGQRIGKGGDSRSDLMSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMFADRETVYAWFTFSHWLVYANSAANPIIYNFLSGKFREEFKAAFSC----------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.25 | 0.24 | 0.71 | 2.88 | Download | ----------------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL---------------------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.23 | 0.20 | 0.73 | 1.70 | Download | -------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE-DVDVIECSLQFPDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEMEGLRTNGVITKRQQKRTGTWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------------------------------------------ | |||||||||||||||||||
| 8 | 4djhA | 0.24 | 0.20 | 0.73 | 4.44 | Download | ----------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDV-IECSLQFPDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYNQTPNRAKRVITTFRTGTWDAYREK-----DRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------------- | |||||||||||||||||||
| 9 | 5vblB | 0.20 | 0.21 | 0.78 | 2.65 | Download | -----------------------------------------CEYTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLEN-TNKVQCYMDYSMWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKNPEDGDPDNGVNPGTDIPDDWVCPLCGVGKDQFEEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWPLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.25 | 0.21 | 0.70 | 2.57 | Download | -----------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQG---SIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLK---------------------------SVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETT-FQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
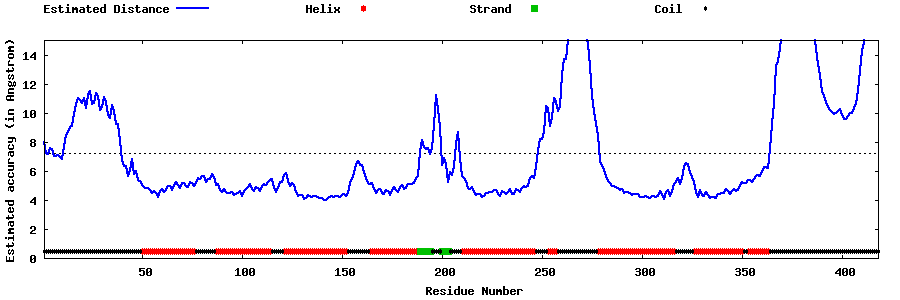
| Generated 3D models | Estimated local accuracy of models | ||
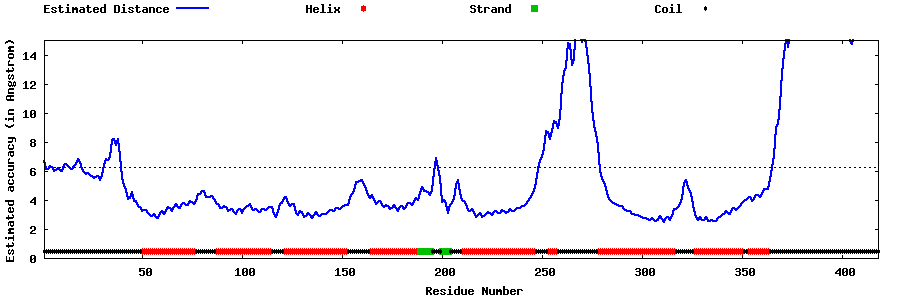 | |||
|
|
|||
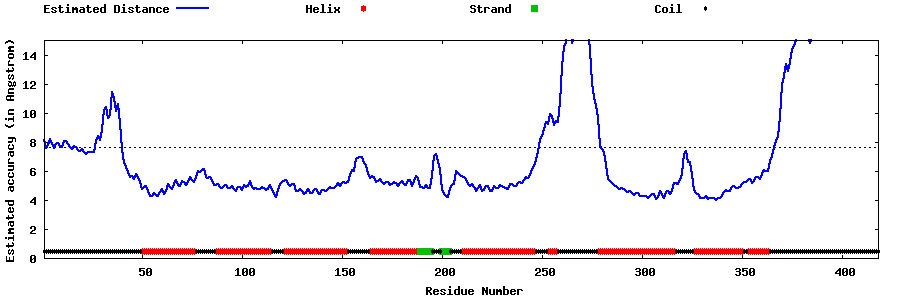 | |||
|
| |||
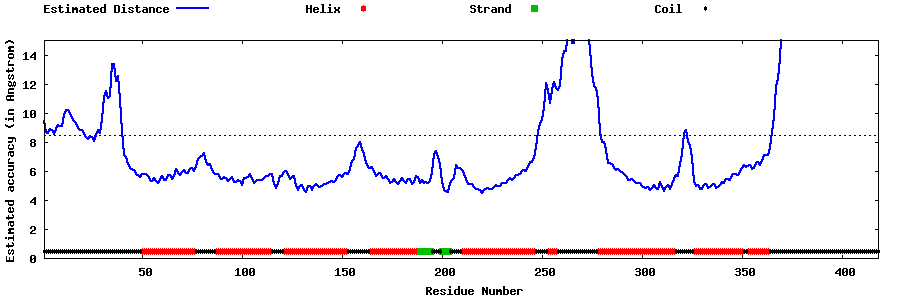 | |||
|
| |||
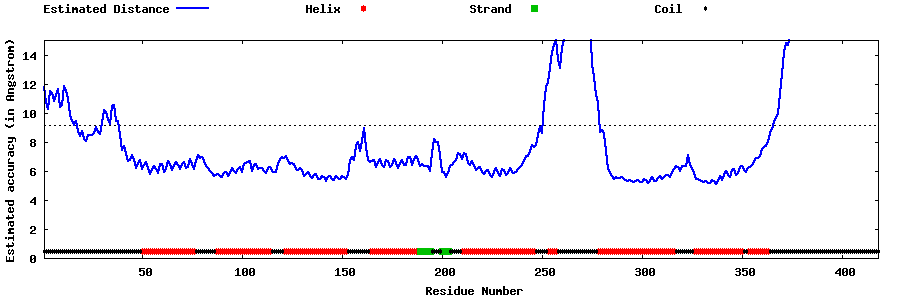 | |||
|
| |||
 | |||
|
| |||