

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MDMLHPSSVSTTSEPENASSAWPPDATLGNVSAGPSPAGLAVSGVLIPLVYLVVCVVGLLGNSLVIYVVLRHTASPSVTNVYILNLALADELFMLGLPFLAAQNALSYWPFGSLMCRLVMAVDGINQFTSIFCLTVMSVDRYLAVVHPTRSARWRTAPVARTVSAAVWVASAVVVLPVVVFSGVPRGMSTCHMQWPEPAAAWRAGFIIYTAALGFFGPLLVICLCYLLIVVKVRSAGRRVWAPSCQRRRRSERRVTRMVVAVVALFVLCWMPFYVLNIVNVVCPLPEEPAFFGLYFLVVALPYANSCANPILYGFLSYRFKQGFRRVLLRPSRRVRSQEPTVGPPEKTEEEDEEEEDGEESREGGKGKEMNGRVSQITQPGTSGQERPPSRVASKEQQLLPQEASTGEKSSTMRISYL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCC | |
| 9987877676778887666777887666776667774422189988999999999999877986413210088889799999999999999999958999999983898786768889999999999999999999999848764300665545467778888689999999999899986798679689975489706689999999999999999999999999999999825578876405666200055154657999999999837999999999984343117999999999999999998999999981898999999995123147666788878765665665666787766568898887887555658988888889755444335656777874013677510059 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MDMLHPSSVSTTSEPENASSAWPPDATLGNVSAGPSPAGLAVSGVLIPLVYLVVCVVGLLGNSLVIYVVLRHTASPSVTNVYILNLALADELFMLGLPFLAAQNALSYWPFGSLMCRLVMAVDGINQFTSIFCLTVMSVDRYLAVVHPTRSARWRTAPVARTVSAAVWVASAVVVLPVVVFSGVPRGMSTCHMQWPEPAAAWRAGFIIYTAALGFFGPLLVICLCYLLIVVKVRSAGRRVWAPSCQRRRRSERRVTRMVVAVVALFVLCWMPFYVLNIVNVVCPLPEEPAFFGLYFLVVALPYANSCANPILYGFLSYRFKQGFRRVLLRPSRRVRSQEPTVGPPEKTEEEDEEEEDGEESREGGKGKEMNGRVSQITQPGTSGQERPPSRVASKEQQLLPQEASTGEKSSTMRISYL | |
| 3342333212223332120321333222333233344322200000002101200220231020000000103321000000000001001100000000000002320100100000000013000100000000002000000000020232112000000000011000000000000020354321010202344320200010010110123112000200000011013234544444344444422000000000000000001100000001001303322000001010000002000200000000043004101300221043244543434444434444444444444544454443444344234344444533345344454431346454454433131315 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCC MDMLHPSSVSTTSEPENASSAWPPDATLGNVSAGPSPAGLAVSGVLIPLVYLVVCVVGLLGNSLVIYVVLRHTASPSVTNVYILNLALADELFMLGLPFLAAQNALSYWPFGSLMCRLVMAVDGINQFTSIFCLTVMSVDRYLAVVHPTRSARWRTAPVARTVSAAVWVASAVVVLPVVVFSGVPRGMSTCHMQWPEPAAAWRAGFIIYTAALGFFGPLLVICLCYLLIVVKVRSAGRRVWAPSCQRRRRSERRVTRMVVAVVALFVLCWMPFYVLNIVNVVCPLPEEPAFFGLYFLVVALPYANSCANPILYGFLSYRFKQGFRRVLLRPSRRVRSQEPTVGPPEKTEEEDEEEEDGEESREGGKGKEMNGRVSQITQPGTSGQERPPSRVASKEQQLLPQEASTGEKSSTMRISYL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.40 | 0.29 | 0.71 | 3.14 | Download | -------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP--------------------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.39 | 0.31 | 0.71 | 3.82 | Download | ------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRS---LRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK---------------------------------------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.39 | 0.29 | 0.72 | 3.85 | Download | -------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.38 | 0.29 | 0.68 | 1.57 | Download | ---------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIPYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.38 | 0.29 | 0.68 | 1.18 | Download | ---------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNTPYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA-----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.39 | 0.29 | 0.71 | 3.39 | Download | ---------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.27 | 0.21 | 0.67 | 1.73 | Download | -------------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVEDGECYIQFF-----SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGADAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCI-P--NTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.43 | 0.30 | 0.66 | 5.01 | Download | ------------------------------------P---LGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLTLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHP-------TSSKAQAVNVAIWALASVVGVPVAIMGSAQDEEIECLVEIPTPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS---REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR--------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.39 | 0.29 | 0.72 | 3.19 | Download | -------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRS---LRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.38 | 0.27 | 0.70 | 3.60 | Download | ----------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS---KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------------------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
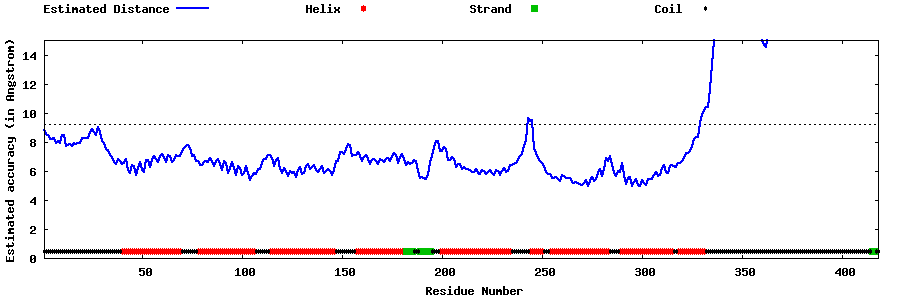
| Generated 3D models | Estimated local accuracy of models | ||
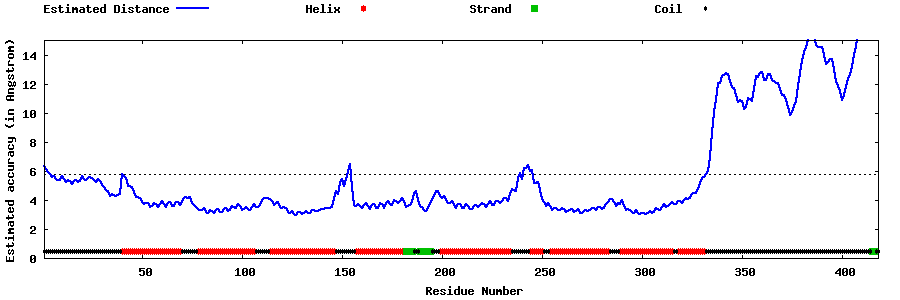 | |||
|
|
|||
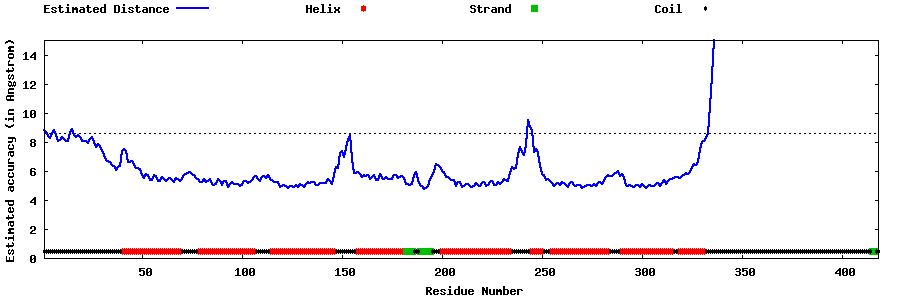 | |||
|
| |||
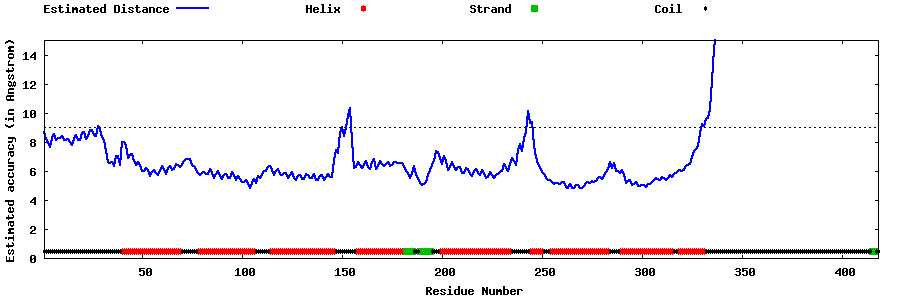 | |||
|
| |||
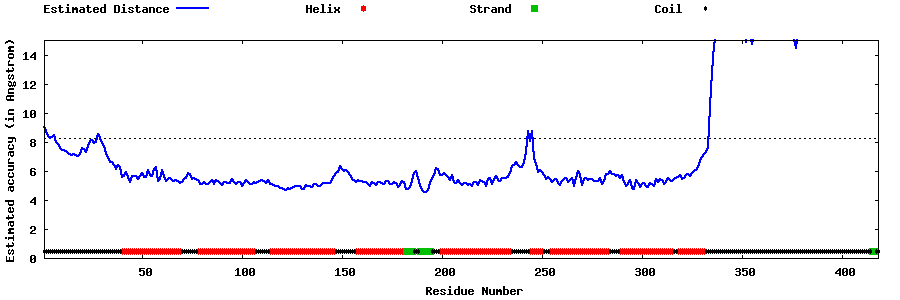 | |||
|
| |||
 | |||
|
| |||