

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNYPLTLEMDLENLEDLFWELDRLDNYNDTSLVENHLCPATEGPLMASFKAVFVPVAYSLIFLLGVIGNVLVLVILERHRQTRSSTETFLFHLAVADLLLVFILPFAVAEGSVGWVLGTFLCKTVIALHKVNFYCSSLLLACIAVDRYLAIVHAVHAYRHRRLLSIHITCGTIWLVGFLLALPEILFAKVSQGHHNNSLPRCTFSQENQAETHAWFTSRFLYHVAGFLLPMLVMGWCYVGVVHRLRQAQRRPQRQKAVRVAILVTSIFFLCWSPYHIVIFLDTLARLKAVDNTCKLNGSLPVAITMCEFLGLAHCCLNPMLYTFAGVKFRSDLSRLLTKLGCTGPASLCQLFPSWRRSSLSESENATSLTTF | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 999876777777676666676666778777766677787544410362899879999999999999889997786662788786999999999999999999699999998479978578899999999999999999999999997899987353243665036777579999999999999999862366169982489841689721588999999999999999999999999999999984266885454479999999999999978999999999999844678866899999999999999999999688999996188899999999874378882224567888887887899998787879 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNYPLTLEMDLENLEDLFWELDRLDNYNDTSLVENHLCPATEGPLMASFKAVFVPVAYSLIFLLGVIGNVLVLVILERHRQTRSSTETFLFHLAVADLLLVFILPFAVAEGSVGWVLGTFLCKTVIALHKVNFYCSSLLLACIAVDRYLAIVHAVHAYRHRRLLSIHITCGTIWLVGFLLALPEILFAKVSQGHHNNSLPRCTFSQENQAETHAWFTSRFLYHVAGFLLPMLVMGWCYVGVVHRLRQAQRRPQRQKAVRVAILVTSIFFLCWSPYHIVIFLDTLARLKAVDNTCKLNGSLPVAITMCEFLGLAHCCLNPMLYTFAGVKFRSDLSRLLTKLGCTGPASLCQLFPSWRRSSLSESENATSLTTF | |
| 874424240434423322233333332332323434224224443143010000021012002303333220010001123311000000000010111000000000000021010111011101001221110000000000310200000003023223211000000000200000010000002024356543111010303563332010011111111012211210220000000101316644422000000000000000031100000001002304113440323300100011010000200021000000004400410130034213344543363344444444344744434336 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MNYPLTLEMDLENLEDLFWELDRLDNYNDTSLVENHLCPATEGPLMASFKAVFVPVAYSLIFLLGVIGNVLVLVILERHRQTRSSTETFLFHLAVADLLLVFILPFAVAEGSVGWVLGTFLCKTVIALHKVNFYCSSLLLACIAVDRYLAIVHAVHAYRHRRLLSIHITCGTIWLVGFLLALPEILFAKVSQGHHNNSLPRCTFSQENQAETHAWFTSRFLYHVAGFLLPMLVMGWCYVGVVHRLRQAQRRPQRQKAVRVAILVTSIFFLCWSPYHIVIFLDTLARLKAVDNTCKLNGSLPVAITMCEFLGLAHCCLNPMLYTFAGVKFRSDLSRLLTKLGCTGPASLCQLFPSWRRSSLSESENATSLTTF | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.29 | 0.26 | 0.79 | 3.34 | Download | ---------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL---HYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGL-NNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.29 | 0.26 | 0.79 | 4.36 | Download | ----------------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL---HYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.29 | 0.26 | 0.79 | 3.79 | Download | ---------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLH---YTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN-CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.26 | 0.23 | 0.75 | 1.55 | Download | ----------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKV-REDVDVIECSLQ-FPDDDYSWWLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.25 | 0.23 | 0.75 | 1.18 | Download | ----------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRED-VDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALG---SA---------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.29 | 0.26 | 0.79 | 3.38 | Download | ----------------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL---HYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.26 | 0.23 | 0.74 | 1.71 | Download | -------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRED-VDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-----A-------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF---------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.29 | 0.26 | 0.78 | 5.01 | Download | ---------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL---HYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREKKRHRD---VRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGL-NNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.29 | 0.26 | 0.79 | 3.62 | Download | ---------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKE---GLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ---------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.26 | 0.24 | 0.77 | 4.05 | Download | -----------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR---QGSIDCTLTFSHP-TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
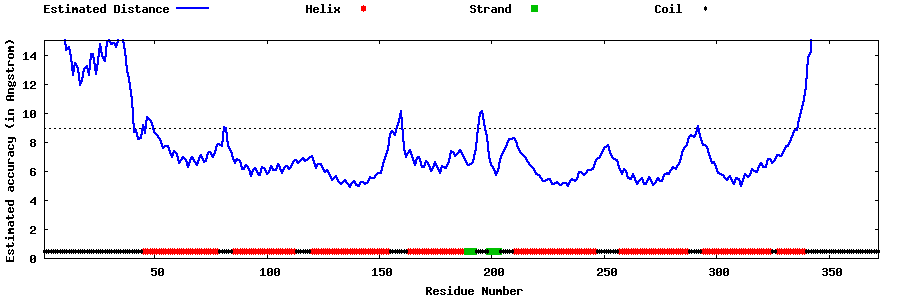
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||