

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDIQMANNFTPPSATPQGNDCDLYAHHSTARIVMPLHYSLVFIIGLVGNLLALVVIVQNRKKINSTTLYSTNLVISDILFTTALPTRIAYYAMGFDWRIGDALCRITALVFYINTYAGVNFMTCLSIDRFIAVVHPLRYNKIKRIEHAKGVCIFVWILVFAQTLPLLINPMSKQEAERITCMEYPNFEETKSLPWILLGACFIGYVLPLIIILICYSQICCKLFRTAKQNPLTEKSGVNKKALNTIILIIVVFVLCFTPYHVAIIQHMIKKLRFSNFLECSQRHSFQISLHFTVCLMNFNCCMDPFIYFFACKGYKRKVMRMLKRQVSVSISSAVKSAPEENSREMTETQMMIHSKSSNGK | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9887778888988898898776655242899999999999999999999998897651578786889999999999999999889999998469987678688999999999999999999999999815887511215546551478899999999999999999985035856997799845997317899999999999999999999999999999998323678873010000014899999999999998368999999999998635778883699999999999999999999998999992388899999999862045765566787888998887777777888888898 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDIQMANNFTPPSATPQGNDCDLYAHHSTARIVMPLHYSLVFIIGLVGNLLALVVIVQNRKKINSTTLYSTNLVISDILFTTALPTRIAYYAMGFDWRIGDALCRITALVFYINTYAGVNFMTCLSIDRFIAVVHPLRYNKIKRIEHAKGVCIFVWILVFAQTLPLLINPMSKQEAERITCMEYPNFEETKSLPWILLGACFIGYVLPLIIILICYSQICCKLFRTAKQNPLTEKSGVNKKALNTIILIIVVFVLCFTPYHVAIIQHMIKKLRFSNFLECSQRHSFQISLHFTVCLMNFNCCMDPFIYFFACKGYKRKVMRMLKRQVSVSISSAVKSAPEENSREMTETQMMIHSKSSNGK | |
| 7436344322333334444414343344011000022012002303323220010001223433101000000000000000000010000034443100300120001111311100020000003100100010020242332200000000001001000101000220355732100000154732201000111211301321221002000100010020345444444444210000000000000000123101000010023032243340423300200110010000300021000000004600410140034223333453354444443443343444444545658 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDIQMANNFTPPSATPQGNDCDLYAHHSTARIVMPLHYSLVFIIGLVGNLLALVVIVQNRKKINSTTLYSTNLVISDILFTTALPTRIAYYAMGFDWRIGDALCRITALVFYINTYAGVNFMTCLSIDRFIAVVHPLRYNKIKRIEHAKGVCIFVWILVFAQTLPLLINPMSKQEAERITCMEYPNFEETKSLPWILLGACFIGYVLPLIIILICYSQICCKLFRTAKQNPLTEKSGVNKKALNTIILIIVVFVLCFTPYHVAIIQHMIKKLRFSNFLECSQRHSFQISLHFTVCLMNFNCCMDPFIYFFACKGYKRKVMRMLKRQVSVSISSAVKSAPEENSREMTETQMMIHSKSSNGK | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.26 | 0.23 | 0.82 | 3.32 | Download | -------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQ--EFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.26 | 0.25 | 0.83 | 4.30 | Download | -------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKYTEEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEF--FGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.24 | 0.22 | 0.84 | 3.74 | Download | ------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-------DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.26 | 0.23 | 0.79 | 1.54 | Download | --------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTVEDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALG---SA----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------ | |||||||||||||||||||
| 5 | 4xnv | 0.27 | 0.25 | 0.83 | 1.16 | Download | -----------------SFKCAL-TKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYTGEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQTPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.24 | 0.22 | 0.83 | 3.41 | Download | --------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI-------DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.17 | 0.22 | 0.80 | 1.68 | Download | -------------------------VPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFWGAYTLEGVLCNCSFDYISRDS-TTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEW----------VTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWV----------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.24 | 0.22 | 0.84 | 4.75 | Download | -----------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.26 | 0.23 | 0.82 | 3.54 | Download | -------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKE------EEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQ--EFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.26 | 0.25 | 0.82 | 4.60 | Download | ---------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKAL--------ITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
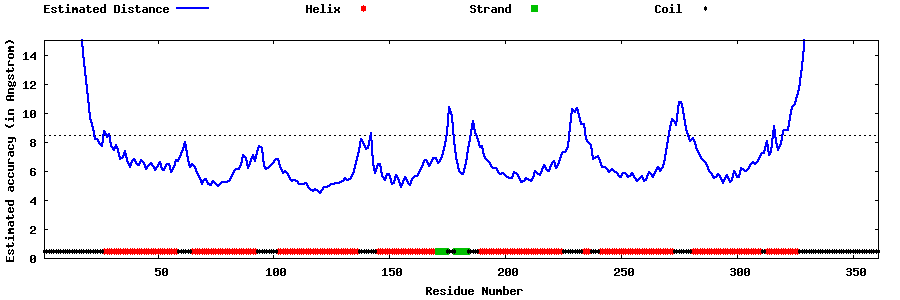
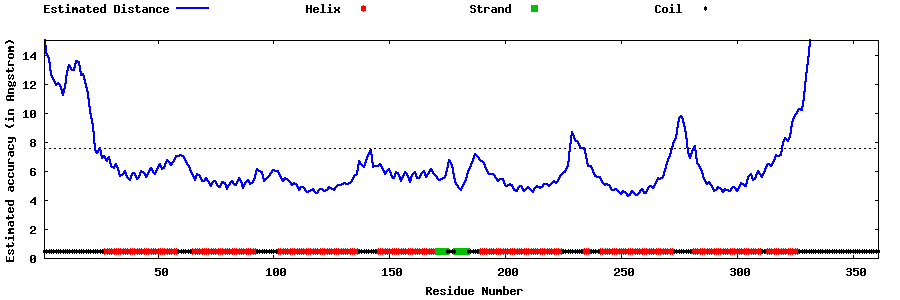
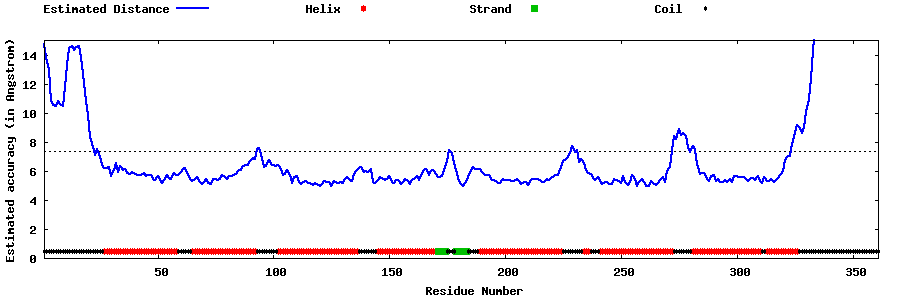
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||