

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDLGKPMKSVLVVALLVIFQVCLCQDEVTDDYIGDNTTVDYTLFESLCSKKDVRNFKAWFLPIMYSIICFVGLLGNGLVVLTYIYFKRLKTMTDTYLLNLAVADILFLLTLPFWAYSAAKSWVFGVHFCKLIFAIYKMSFFSGMLLLLCISIDRYVAIVQAVSAHRHRARVLLISKLSCVGIWILATVLSIPELLYSDLQRSSSEQAMRCSLITEHVEAFITIQVAQMVIGFLVPLLAMSFCYLVIIRTLLQARNFERNKAIKVIIAVVVVFIVFQLPYNGVVLAQTVANFNITSSTCELSKQLNIAYDVTYSLACVRCCVNPFLYAFIGVKFRNDLFKLFKDLGCLSQEQLRQWSSCRHIRRSSMSVEAETTTTFSP | |
| CCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 998884288899999998454037788876666787876887676778632248789988999999999999988998678777287768699999999999999999989999999468997777889999999999999999999999998368876415223256841276999999999999999989999751377589867998368970779999999999999999999999999999999842688667617999999999999997899999999999983678887159999999999999999999989899999408889999999974346775134567788888888988888998888997 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDLGKPMKSVLVVALLVIFQVCLCQDEVTDDYIGDNTTVDYTLFESLCSKKDVRNFKAWFLPIMYSIICFVGLLGNGLVVLTYIYFKRLKTMTDTYLLNLAVADILFLLTLPFWAYSAAKSWVFGVHFCKLIFAIYKMSFFSGMLLLLCISIDRYVAIVQAVSAHRHRARVLLISKLSCVGIWILATVLSIPELLYSDLQRSSSEQAMRCSLITEHVEAFITIQVAQMVIGFLVPLLAMSFCYLVIIRTLLQARNFERNKAIKVIIAVVVVFIVFQLPYNGVVLAQTVANFNITSSTCELSKQLNIAYDVTYSLACVRCCVNPFLYAFIGVKFRNDLFKLFKDLGCLSQEQLRQWSSCRHIRRSSMSVEAETTTTFSP | |
| 853734234020210220010003333233313342332314335320436314400000002001200120232321000000102331000000000001000000000000000002001003001110100122111000000000031010000001302422331011000000001100000010000002024476421010103263320110011121110232102102200000001023154453300000000000001001220000000100231312344142230010000001000120002100000000440141014002410323445344444464343444446454444447 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDLGKPMKSVLVVALLVIFQVCLCQDEVTDDYIGDNTTVDYTLFESLCSKKDVRNFKAWFLPIMYSIICFVGLLGNGLVVLTYIYFKRLKTMTDTYLLNLAVADILFLLTLPFWAYSAAKSWVFGVHFCKLIFAIYKMSFFSGMLLLLCISIDRYVAIVQAVSAHRHRARVLLISKLSCVGIWILATVLSIPELLYSDLQRSSSEQAMRCSLITEHVEAFITIQVAQMVIGFLVPLLAMSFCYLVIIRTLLQARNFERNKAIKVIIAVVVVFIVFQLPYNGVVLAQTVANFNITSSTCELSKQLNIAYDVTYSLACVRCCVNPFLYAFIGVKFRNDLFKLFKDLGCLSQEQLRQWSSCRHIRRSSMSVEAETTTTFSP | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.37 | 0.30 | 0.78 | 3.12 | Download | ----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVT--FGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------ | |||||||||||||||||||
| 2 | 4mbsA | 0.37 | 0.29 | 0.78 | 4.30 | Download | ----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKART--VTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------ | |||||||||||||||||||
| 3 | 4mbsA | 0.37 | 0.30 | 0.78 | 3.84 | Download | ----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTV--TFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN-CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------ | |||||||||||||||||||
| 4 | 4djh | 0.26 | 0.25 | 0.73 | 1.53 | Download | -----------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFR--TPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.38 | 0.30 | 0.75 | 1.17 | Download | ----------------------------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAANEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVT--FGVVTSVITWLVAVFASVPGIIFTKQKE-D--SVYVCGPYFPRG-WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRIPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFG-LSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.37 | 0.30 | 0.77 | 3.25 | Download | -----------------------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKART--VTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------ | |||||||||||||||||||
| 7 | 5t1a | 0.38 | 0.30 | 0.74 | 1.72 | Download | ------------------------------------------------------QIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAANEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTF--GVVTSVITWLVAVFASVPGIIFTKQKED---SVYVCGPYFPRG-WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRIPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFG-LSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.37 | 0.29 | 0.77 | 4.86 | Download | ----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKART--VTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK--KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN-CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------ | |||||||||||||||||||
| 9 | 4mbsA | 0.37 | 0.30 | 0.78 | 3.51 | Download | ----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKART--VTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.27 | 0.22 | 0.76 | 3.79 | Download | ------------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRT--PRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSLRRITRMVLVVVAVFIVCWTPIHIYVIIKAL-------ITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
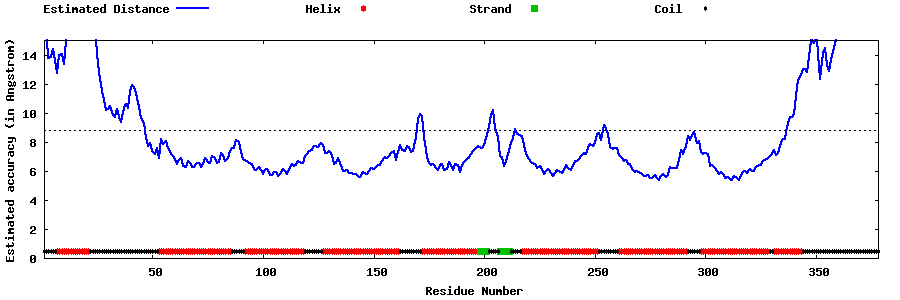
| Generated 3D models | Estimated local accuracy of models | ||
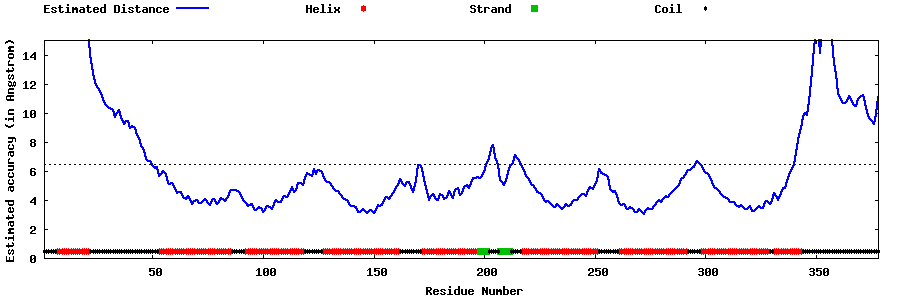 | |||
|
|
|||
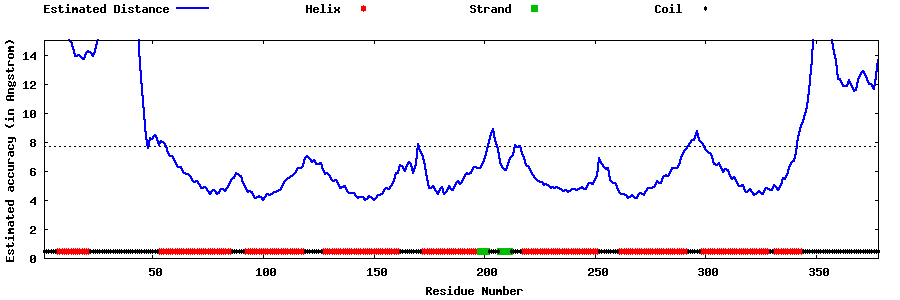 | |||
|
| |||
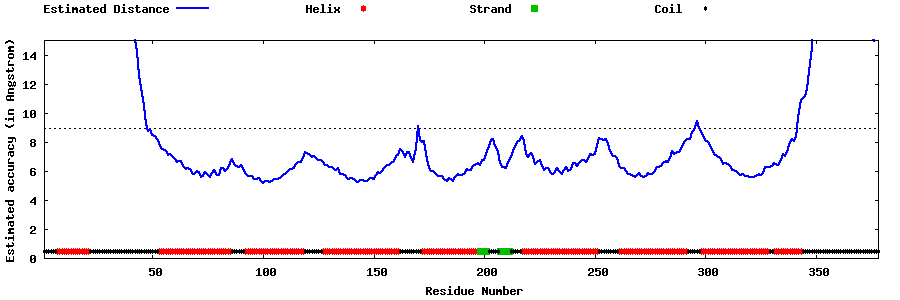 | |||
|
| |||
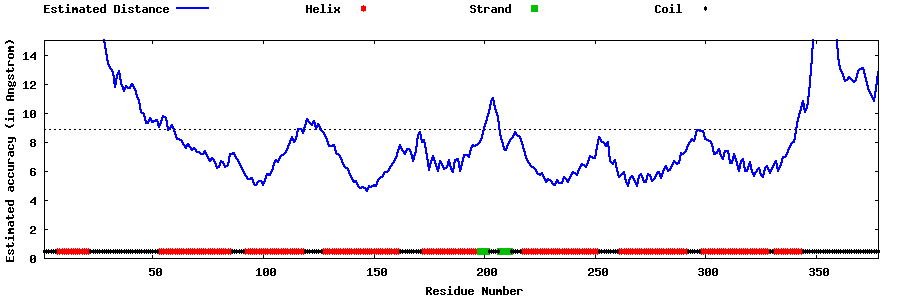 | |||
|
| |||
 | |||
|
| |||