

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MANSASPEQNQNHCSAINNSIPLMQGNLPTLTLSGKIRVTVTFFLFLLSATFNASFLLKLQKWTQKKEKGKKLSRMKLLLKHLTLANLLETLIVMPLDGMWNITVQWYAGELLCKVLSYLKLFSMYAPAFMMVVISLDRSLAITRPLALKSNSKVGQSMVGLAWILSSVFAGPQLYIFRMIHLADSSGQTKVFSQCVTHCSFSQWWHQAFYNFFTFSCLFIIPLFIMLICNAKIIFTLTRVLHQDPHELQLNQSKNNIPRARLKTLKMTVAFATSFTVCWTPYYVLGIWYWFDPEMLNRLSDPVNHFFFLFAFLNPCFDPLIYGYFSL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCC | |
| 9999999767776666788887676778776557999999999999999997887622300101213477788749999999999999999997379999987060534158997899999999999999999999986999764787665422688878899999999999999962667623788761001342258887645899999999999999999999999999999999986067888632011332888998799999999999999997999999999996743143058999999999999999599999983279 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MANSASPEQNQNHCSAINNSIPLMQGNLPTLTLSGKIRVTVTFFLFLLSATFNASFLLKLQKWTQKKEKGKKLSRMKLLLKHLTLANLLETLIVMPLDGMWNITVQWYAGELLCKVLSYLKLFSMYAPAFMMVVISLDRSLAITRPLALKSNSKVGQSMVGLAWILSSVFAGPQLYIFRMIHLADSSGQTKVFSQCVTHCSFSQWWHQAFYNFFTFSCLFIIPLFIMLICNAKIIFTLTRVLHQDPHELQLNQSKNNIPRARLKTLKMTVAFATSFTVCWTPYYVLGIWYWFDPEMLNRLSDPVNHFFFLFAFLNPCFDPLIYGYFSL | |
| 4513233433433232223323236442331321020102011200220131120000000112313445433000000020003001210200020200010133020030001003101010000001000000000020001002223343001000000012001101000000102324436332300120013021434103100000103231331021002001200010133245354544444444424312100000000000010113231100000100143124302200100001001210013031303124 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCC MANSASPEQNQNHCSAINNSIPLMQGNLPTLTLSGKIRVTVTFFLFLLSATFNASFLLKLQKWTQKKEKGKKLSRMKLLLKHLTLANLLETLIVMPLDGMWNITVQWYAGELLCKVLSYLKLFSMYAPAFMMVVISLDRSLAITRPLALKSNSKVGQSMVGLAWILSSVFAGPQLYIFRMIHLADSSGQTKVFSQCVTHCSFSQWWHQAFYNFFTFSCLFIIPLFIMLICNAKIIFTLTRVLHQDPHELQLNQSKNNIPRARLKTLKMTVAFATSFTVCWTPYYVLGIWYWFDPEMLNRLSDPVNHFFFLFAFLNPCFDPLIYGYFSL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.17 | 0.18 | 0.86 | 2.60 | Download | ----------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK------TATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-------AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS--------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDE | |||||||||||||||||||
| 2 | 1gzmA | 0.15 | 0.20 | 0.94 | 3.79 | Download | MNGTEGPNFPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLR------TPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEG-----MQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQES-------ATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQG--SDFGPIFMTIPAFFAKTSAVYNPVIYIMMN- | |||||||||||||||||||
| 3 | 4n6hA | 0.17 | 0.18 | 0.87 | 3.54 | Download | ----------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK------TATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-------AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSK--------EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDE | |||||||||||||||||||
| 4 | 4ib4 | 0.18 | 0.23 | 0.86 | 1.54 | Download | ----------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSL--EK----KLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYRATAFIKITVVWLISIGIAIPVPIKGI-ETN-------PNNITCV----LT-KERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWEAIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNK | |||||||||||||||||||
| 5 | 4djh | 0.19 | 0.23 | 0.86 | 1.16 | Download | ------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMK------TATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVD-----VIECSLQFPDDDSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIVIQTPAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDE | |||||||||||||||||||
| 6 | 4n6hA | 0.17 | 0.18 | 0.86 | 2.96 | Download | ------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK------TATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-------AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV--------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDE | |||||||||||||||||||
| 7 | 3uon | 0.20 | 0.23 | 0.83 | 1.75 | Download | ------------------------------------FIVLVAGSLSLVTIIGNILVMVSIKVNRHLQ------TVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDG----ECYIQFF-S----NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEGGLAATWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP---CIPNTVWTIGYWLCYINSTINPACYALCN- | |||||||||||||||||||
| 8 | 4djhA | 0.18 | 0.23 | 0.86 | 3.43 | Download | ------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMK------TATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDV----IECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEMLRIDEEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA-----LSSYYFCIALGYTNSSLNPILYAFLDE | |||||||||||||||||||
| 9 | 4grvA | 0.20 | 0.23 | 0.86 | 2.67 | Download | ----------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR-KSLQ------STVHYHLGSLALSDLLILLLAMPVELYNFIWHPWAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAICHPFKAKMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTH----PGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMSGSV--------------QALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQWTFYHYFYMLTNALAYASSAINPILYNLVSV | |||||||||||||||||||
| 10 | 5c1mA | 0.20 | 0.21 | 0.88 | 3.60 | Download | ------------------GSHSLPQTGSPSM-VTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMK------TATNIYIFNLALADALATS-TLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKAFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGS-------IDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLK--------SVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDE | |||||||||||||||||||
| ||||||||||||||||||||||||||
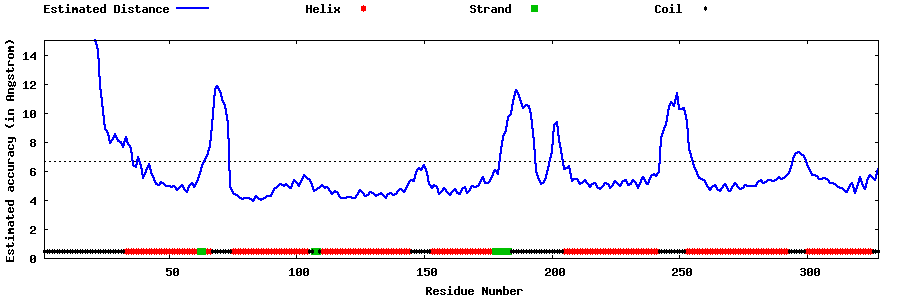
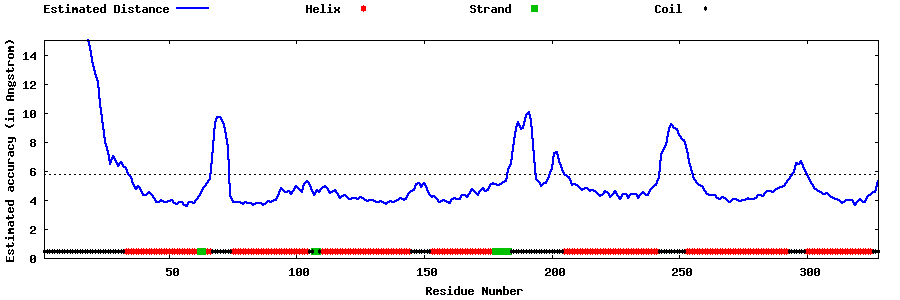
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||