

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDMADEPLNGSHTWLSIPFDLNGSVVSTNTSNQTEPYYDLTSNAVLTFIYFVVCIIGLCGNTLVIYVILRYAKMKTITNIYILNLAIADELFMLGLPFLAMQVALVHWPFGKAICRVVMTVDGINQFTSIFCLTVMSIDRYLAVVHPIKSAKWRRPRTAKMITMAVWGVSLLVILPIMIYAGLRSNQWGRSSCTINWPGESGAWYTGFIIYTFILGFLVPLTIICLCYLFIIIKVKSSGIRVGSSKRKKSEKKVTRMVSIVVAVFIFCWLPFYIFNVSSVSMAISPTPALKGMFDFVVVLTYANSCANPILYAFLSDNFKKSFQNVLCLVKVSGTDDGERSDSKQDKSRLNETTETQRTLLNGDLQTSI | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 999888888776666778788877677888876564420488999999999999999876885010013088789799999999999999999968999999973898897678889999999999999999999999858887500653326467889999899999999999899987166887899679973799715689999999999999999999999999999999817788876420110024053235999999999978999999999970267318999999999999999999999999980999999999983742368877666677878777778888652016788838889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDMADEPLNGSHTWLSIPFDLNGSVVSTNTSNQTEPYYDLTSNAVLTFIYFVVCIIGLCGNTLVIYVILRYAKMKTITNIYILNLAIADELFMLGLPFLAMQVALVHWPFGKAICRVVMTVDGINQFTSIFCLTVMSIDRYLAVVHPIKSAKWRRPRTAKMITMAVWGVSLLVILPIMIYAGLRSNQWGRSSCTINWPGESGAWYTGFIIYTFILGFLVPLTIICLCYLFIIIKVKSSGIRVGSSKRKKSEKKVTRMVSIVVAVFIFCWLPFYIFNVSSVSMAISPTPALKGMFDFVVVLTYANSCANPILYAFLSDNFKKSFQNVLCLVKVSGTDDGERSDSKQDKSRLNETTETQRTLLNGDLQTSI | |
| 663544423333322323232221233333333443223200100002211200230231121001000103320100000000001021201000001000002420100100000000003110100000000003131000000020232122110000000010000010100000000243743211010302354220210010110231233122000200000011023333444444443432000000000000010031211000001002404333001001010000002000200000000144004101300211323445534444344444444444434444454644347 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDMADEPLNGSHTWLSIPFDLNGSVVSTNTSNQTEPYYDLTSNAVLTFIYFVVCIIGLCGNTLVIYVILRYAKMKTITNIYILNLAIADELFMLGLPFLAMQVALVHWPFGKAICRVVMTVDGINQFTSIFCLTVMSIDRYLAVVHPIKSAKWRRPRTAKMITMAVWGVSLLVILPIMIYAGLRSNQWGRSSCTINWPGESGAWYTGFIIYTFILGFLVPLTIICLCYLFIIIKVKSSGIRVGSSKRKKSEKKVTRMVSIVVAVFIFCWLPFYIFNVSSVSMAISPTPALKGMFDFVVVLTYANSCANPILYAFLSDNFKKSFQNVLCLVKVSGTDDGERSDSKQDKSRLNETTETQRTLLNGDLQTSI | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.40 | 0.33 | 0.82 | 3.40 | Download | ------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.40 | 0.33 | 0.81 | 4.03 | Download | ------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK---------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.39 | 0.33 | 0.82 | 4.04 | Download | ------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.39 | 0.33 | 0.78 | 1.57 | Download | --------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS--A---ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.39 | 0.33 | 0.78 | 1.17 | Download | --------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.39 | 0.33 | 0.81 | 3.53 | Download | --------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.39 | 0.33 | 0.77 | 1.72 | Download | ---------------------------------------PAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF----------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.39 | 0.30 | 0.75 | 5.17 | Download | --------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLTLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHP-------TSSKAQAVNVAIWALASVVGVPVAIMGSAQ-VEDEEIECLVEIPTPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGSREKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR--------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.39 | 0.33 | 0.82 | 3.42 | Download | ------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.41 | 0.33 | 0.80 | 4.44 | Download | ---------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQ-GSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
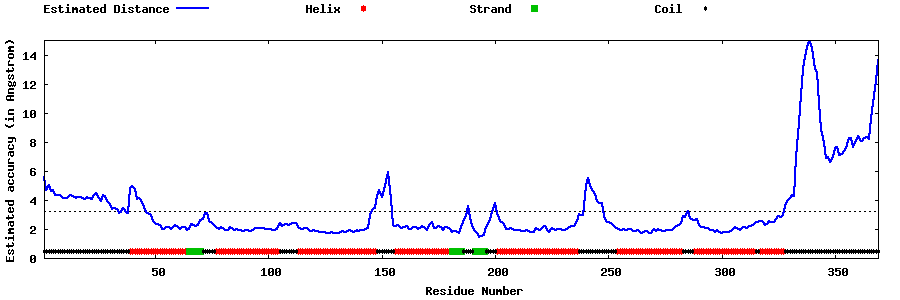 | |||
|
|
|||
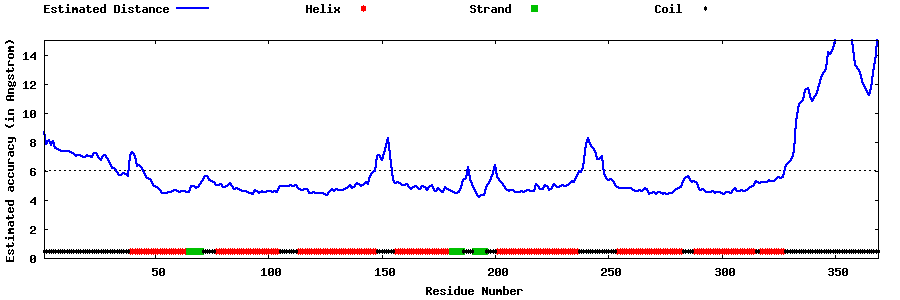 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||