

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MEGALAANWSAEAANASAAPPGAEGNRTAGPPRRNEALARVEVAVLCLILLLALSGNACVLLALRTTRQKHSRLFFFMKHLSIADLVVAVFQVLPQLLWDITFRFYGPDLLCRLVKYLQVVGMFASTYLLLLMSLDRCLAICQPLRSLRRRTDRLAVLATWLGCLVASAPQVHIFSLREVADGVFDCWAVFIQPWGPKAYITWITLAVYIVPVIVLAACYGLISFKIWQNLRLKTAAAAAAEAPEGAAAGDGGRVALARVSSVKLISKAKIRTVKMTFIIVLAFIVCWTPFFFVQMWSVWDANAPKEASAFIIVMLLASLNSCCNPWIYMLFTGHLFHELVQRFLCCSASYLKGRRLGETSASKKSNSSSFVLSHRSSSQRSCSQPSTA | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99877898888888778899998877777887668209999999999999999998997726304279888477999999999999999998899999998293767148988999999999999999999999972887317787641357789999999999999999998578797479860476315883043899999999999999999999999999999774155543024442012333344555441211025578998899999999999999999579999999999677876666999999999999998989999971999999999975777888877677778877765788887766899998888799999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MEGALAANWSAEAANASAAPPGAEGNRTAGPPRRNEALARVEVAVLCLILLLALSGNACVLLALRTTRQKHSRLFFFMKHLSIADLVVAVFQVLPQLLWDITFRFYGPDLLCRLVKYLQVVGMFASTYLLLLMSLDRCLAICQPLRSLRRRTDRLAVLATWLGCLVASAPQVHIFSLREVADGVFDCWAVFIQPWGPKAYITWITLAVYIVPVIVLAACYGLISFKIWQNLRLKTAAAAAAEAPEGAAAGDGGRVALARVSSVKLISKAKIRTVKMTFIIVLAFIVCWTPFFFVQMWSVWDANAPKEASAFIIVMLLASLNSCCNPWIYMLFTGHLFHELVQRFLCCSASYLKGRRLGETSASKKSNSSSFVLSHRSSSQRSCSQPSTA | |
| 62422332132332121233333433334332322320010002111200110130110000000123422000000010002000000000010100000032020010001002001000000000000000000010001002223332000000000000000001000000023345221101010136202100000101110231010002000000010123243454444444344444444444333333344432330110000000000000000021000000010023323221000000010002000000000000043004102410200233335444444433443443333333433344444444638 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MEGALAANWSAEAANASAAPPGAEGNRTAGPPRRNEALARVEVAVLCLILLLALSGNACVLLALRTTRQKHSRLFFFMKHLSIADLVVAVFQVLPQLLWDITFRFYGPDLLCRLVKYLQVVGMFASTYLLLLMSLDRCLAICQPLRSLRRRTDRLAVLATWLGCLVASAPQVHIFSLREVADGVFDCWAVFIQPWGPKAYITWITLAVYIVPVIVLAACYGLISFKIWQNLRLKTAAAAAAEAPEGAAAGDGGRVALARVSSVKLISKAKIRTVKMTFIIVLAFIVCWTPFFFVQMWSVWDANAPKEASAFIIVMLLASLNSCCNPWIYMLFTGHLFHELVQRFLCCSASYLKGRRLGETSASKKSNSSSFVLSHRSSSQRSCSQPSTA | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.22 | 0.23 | 0.75 | 2.53 | Download | --------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---------------------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.18 | 0.22 | 0.83 | 3.88 | Download | -----------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDYKLRICLLHPVQKTYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNIFEMLRIDEGGGSGGDEAEKLFNRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.21 | 0.23 | 0.76 | 3.57 | Download | --------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSK---------------------------EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.14 | 0.16 | 0.93 | 1.51 | Download | ---DLR------DNETWWYNPSIIVHPWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTICFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRL----------N----AKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETED---DKDAETE-IPAGESSDAAPSADAAQM | |||||||||||||||||||
| 5 | 4djh | 0.19 | 0.22 | 0.79 | 1.16 | Download | ----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALTPLKAKIINICIWLLSSSVGISAIVLGGTKVREVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEMLRIDEGRNTNGQKRQTPGTWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.21 | 0.23 | 0.75 | 2.82 | Download | ----------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKARTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL---------------------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.15 | 0.16 | 0.76 | 1.70 | Download | -----------------------------------DAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLN--------------AKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPW------------------------------------------- | |||||||||||||||||||
| 8 | 1u19A | 0.18 | 0.22 | 0.88 | 4.86 | Download | MNGTEGPNFYVPFSNKTGVVRSP--FEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYEETNNESFVIYMFVVHFIIPLIVIFFCYG--------------------------QLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA------------------ | |||||||||||||||||||
| 9 | 5vblB | 0.20 | 0.22 | 0.83 | 2.69 | Download | -----------------------------CEYTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENNKVQCYMDYSMVAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKYTCTVCGYIYNPEDGDIPDDWVCPLCGVGKDQFEEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWPMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR--------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.20 | 0.22 | 0.74 | 2.96 | Download | -----------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATST-LPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR-QGSIDCTLTFSHPYWENLLKICVFIFAFIMPVLIITVCYGLMILRLK---------------------------SVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
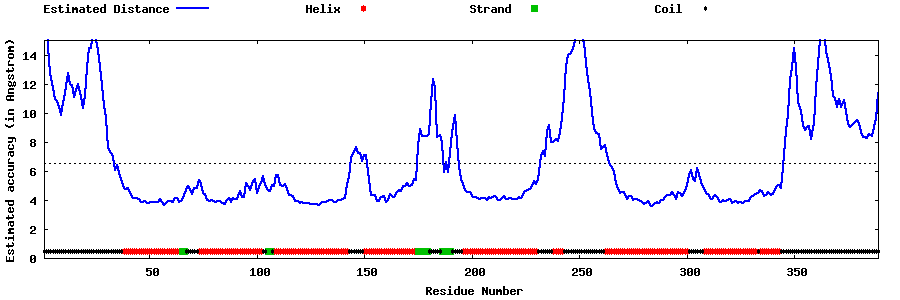
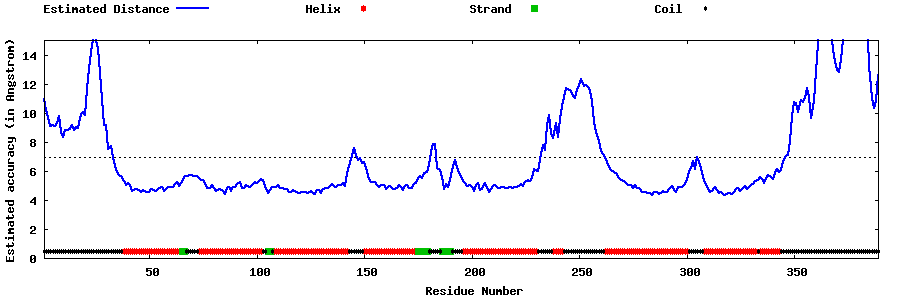
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||